---
license: apache-2.0
datasets:
- jxie/guacamol
- AdrianM0/MUV
library_name: transformers
---
## Model Details
We introduce a suite of neural language model tools for pre-training, fine-tuning SMILES-based molecular language models. Furthermore, we also provide recipes for semi-supervised recipes for fine-tuning these languages in low-data settings using Semi-supervised learning.
### Enumeration-aware Molecular Transformers
Introduces contrastive learning alongside multi-task regression, and masked language modelling as pre-training objectives to inject enumeration knowledge into pre-trained language models.
#### a. Molecular Domain Adaptation (Contrastive Encoder-based)
##### i. Architecture

##### ii. Contrastive Learning
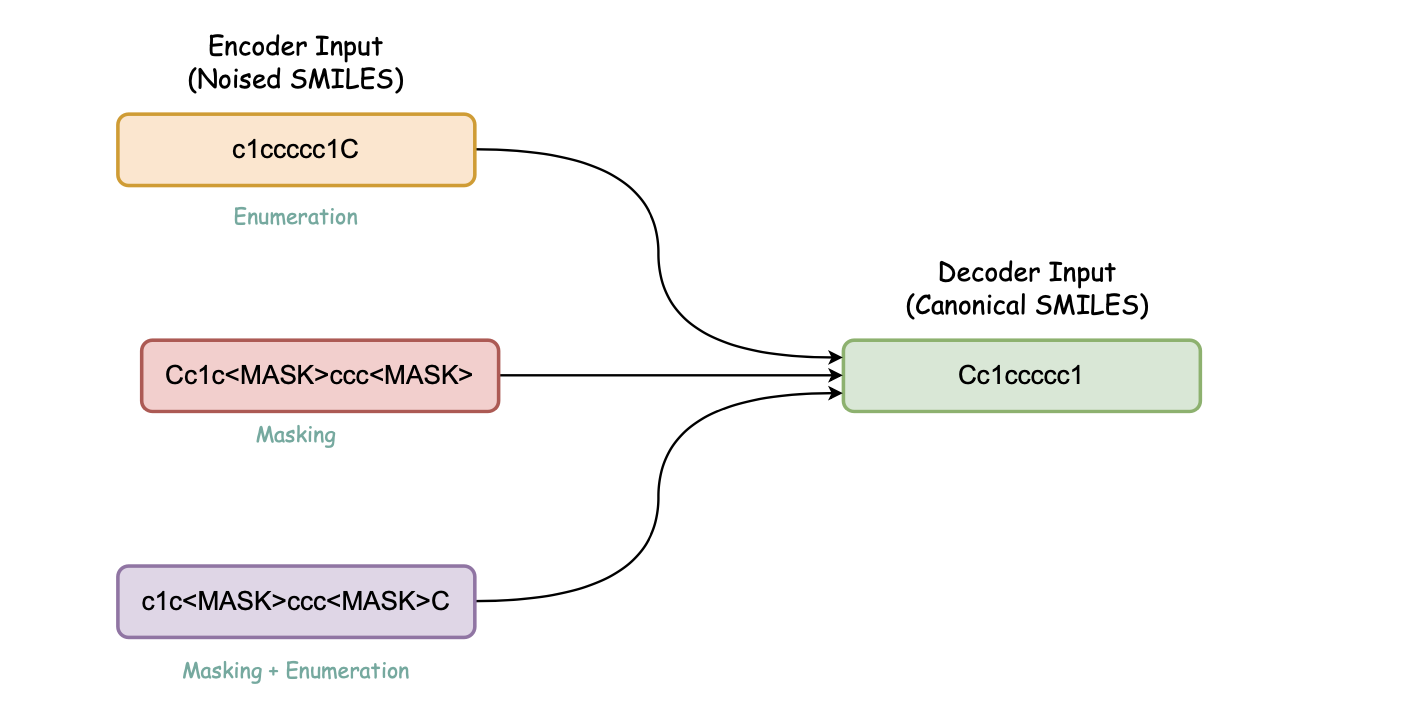
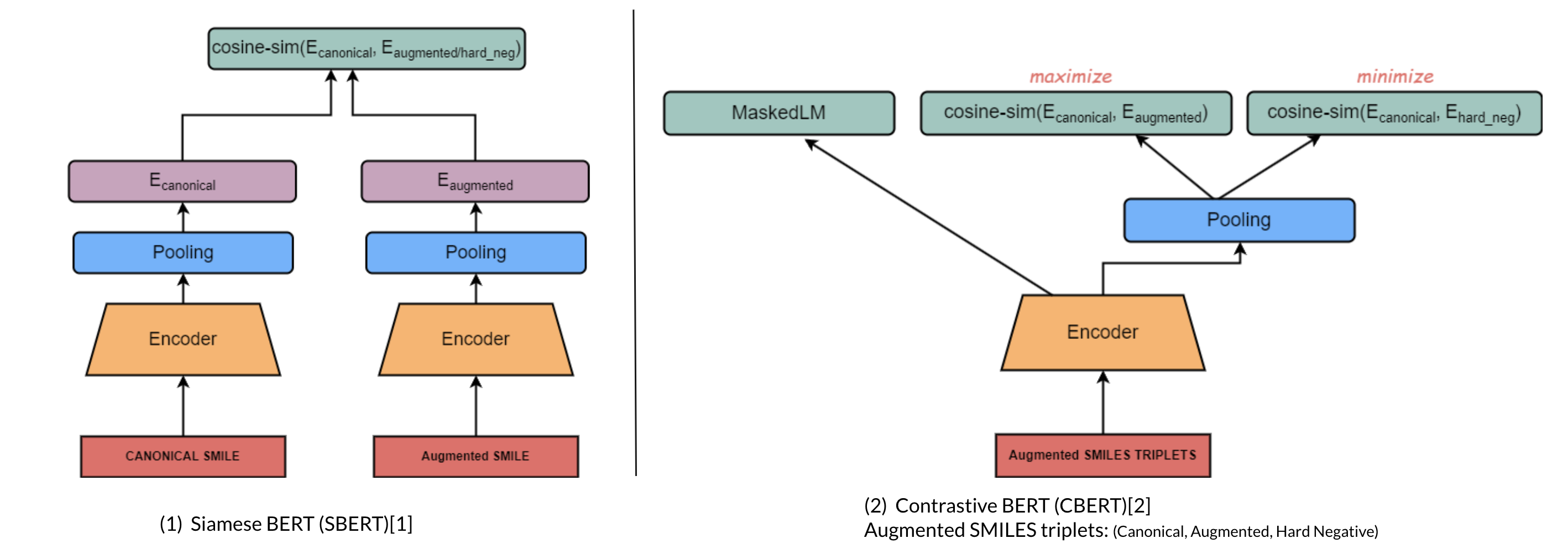 #### b. Canonicalization Encoder-decoder (Denoising Encoder-decoder)
#### b. Canonicalization Encoder-decoder (Denoising Encoder-decoder)
 ### Pretraining steps for this model:
- Pretrain BERT model with Multi task regression on physicochemical properties on Guacamol dataset
- Domain adaptation on MUV dataset with Constrastive Learning, Masked Language Modeling
Fore more details please see our [github repository](https://github.com/uds-lsv/enumeration-aware-molecule-transformers).
### Pretraining steps for this model:
- Pretrain BERT model with Multi task regression on physicochemical properties on Guacamol dataset
- Domain adaptation on MUV dataset with Constrastive Learning, Masked Language Modeling
Fore more details please see our [github repository](https://github.com/uds-lsv/enumeration-aware-molecule-transformers). #### b. Canonicalization Encoder-decoder (Denoising Encoder-decoder)
#### b. Canonicalization Encoder-decoder (Denoising Encoder-decoder)
 ### Pretraining steps for this model:
- Pretrain BERT model with Multi task regression on physicochemical properties on Guacamol dataset
- Domain adaptation on MUV dataset with Constrastive Learning, Masked Language Modeling
Fore more details please see our [github repository](https://github.com/uds-lsv/enumeration-aware-molecule-transformers).
### Pretraining steps for this model:
- Pretrain BERT model with Multi task regression on physicochemical properties on Guacamol dataset
- Domain adaptation on MUV dataset with Constrastive Learning, Masked Language Modeling
Fore more details please see our [github repository](https://github.com/uds-lsv/enumeration-aware-molecule-transformers).