Commit
•
b0fb15f
1
Parent(s):
fe94515
Upload 15 files
Browse files- .gitattributes +2 -35
- LICENSE +21 -0
- README.md +34 -3
- fig/CLS.png +0 -0
- fig/NeuralCGMM.png +0 -0
- fig/allpic.png +0 -0
- fig/output.png +0 -0
- fig/problem.png +0 -0
- model/README.md +9 -0
- model/data/best_model.pt +0 -0
- model/data/test-data.pt +0 -0
- model/data/torch_dist.pt +0 -0
- model/dataloader.py +36 -0
- model/main.py +176 -0
- model/models.md +12 -0
.gitattributes
CHANGED
|
@@ -1,35 +1,2 @@
|
|
| 1 |
-
|
| 2 |
-
|
| 3 |
-
*.bin filter=lfs diff=lfs merge=lfs -text
|
| 4 |
-
*.bz2 filter=lfs diff=lfs merge=lfs -text
|
| 5 |
-
*.ckpt filter=lfs diff=lfs merge=lfs -text
|
| 6 |
-
*.ftz filter=lfs diff=lfs merge=lfs -text
|
| 7 |
-
*.gz filter=lfs diff=lfs merge=lfs -text
|
| 8 |
-
*.h5 filter=lfs diff=lfs merge=lfs -text
|
| 9 |
-
*.joblib filter=lfs diff=lfs merge=lfs -text
|
| 10 |
-
*.lfs.* filter=lfs diff=lfs merge=lfs -text
|
| 11 |
-
*.mlmodel filter=lfs diff=lfs merge=lfs -text
|
| 12 |
-
*.model filter=lfs diff=lfs merge=lfs -text
|
| 13 |
-
*.msgpack filter=lfs diff=lfs merge=lfs -text
|
| 14 |
-
*.npy filter=lfs diff=lfs merge=lfs -text
|
| 15 |
-
*.npz filter=lfs diff=lfs merge=lfs -text
|
| 16 |
-
*.onnx filter=lfs diff=lfs merge=lfs -text
|
| 17 |
-
*.ot filter=lfs diff=lfs merge=lfs -text
|
| 18 |
-
*.parquet filter=lfs diff=lfs merge=lfs -text
|
| 19 |
-
*.pb filter=lfs diff=lfs merge=lfs -text
|
| 20 |
-
*.pickle filter=lfs diff=lfs merge=lfs -text
|
| 21 |
-
*.pkl filter=lfs diff=lfs merge=lfs -text
|
| 22 |
-
*.pt filter=lfs diff=lfs merge=lfs -text
|
| 23 |
-
*.pth filter=lfs diff=lfs merge=lfs -text
|
| 24 |
-
*.rar filter=lfs diff=lfs merge=lfs -text
|
| 25 |
-
*.safetensors filter=lfs diff=lfs merge=lfs -text
|
| 26 |
-
saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
| 27 |
-
*.tar.* filter=lfs diff=lfs merge=lfs -text
|
| 28 |
-
*.tar filter=lfs diff=lfs merge=lfs -text
|
| 29 |
-
*.tflite filter=lfs diff=lfs merge=lfs -text
|
| 30 |
-
*.tgz filter=lfs diff=lfs merge=lfs -text
|
| 31 |
-
*.wasm filter=lfs diff=lfs merge=lfs -text
|
| 32 |
-
*.xz filter=lfs diff=lfs merge=lfs -text
|
| 33 |
-
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
-
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
-
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
| 1 |
+
# Auto detect text files and perform LF normalization
|
| 2 |
+
* text=auto
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
LICENSE
ADDED
|
@@ -0,0 +1,21 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
MIT License
|
| 2 |
+
|
| 3 |
+
Copyright (c) 2024 Azmine Toushik Wasi
|
| 4 |
+
|
| 5 |
+
Permission is hereby granted, free of charge, to any person obtaining a copy
|
| 6 |
+
of this software and associated documentation files (the "Software"), to deal
|
| 7 |
+
in the Software without restriction, including without limitation the rights
|
| 8 |
+
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
|
| 9 |
+
copies of the Software, and to permit persons to whom the Software is
|
| 10 |
+
furnished to do so, subject to the following conditions:
|
| 11 |
+
|
| 12 |
+
The above copyright notice and this permission notice shall be included in all
|
| 13 |
+
copies or substantial portions of the Software.
|
| 14 |
+
|
| 15 |
+
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
|
| 16 |
+
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
|
| 17 |
+
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
|
| 18 |
+
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
|
| 19 |
+
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
|
| 20 |
+
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
|
| 21 |
+
SOFTWARE.
|
README.md
CHANGED
|
@@ -1,3 +1,34 @@
|
|
| 1 |
-
|
| 2 |
-
|
| 3 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# ***NeuralCGMM*: Neural Control System for Continuous Glucose Monitoring and Maintenance**
|
| 2 |
+
- **Authors:** Azmine Toushik Wasi
|
| 3 |
+
- **OpenReview**: https://openreview.net/forum?id=Te4P3Cn54g
|
| 4 |
+
- **arXiv**: https://arxiv.org/abs/2402.13852
|
| 5 |
+
|
| 6 |
+
---
|
| 7 |
+
**Abstract:** Precise glucose level monitoring is critical for people with diabetes to avoid serious complications. While there are several methods for continuous glucose level monitoring, research on maintenance devices is limited. To mitigate the gap, we provide a novel neural control system for continuous glucose monitoring and management that uses differential predictive control, NeuralCGMM. Our approach, led by a sophisticated neural policy and differentiable modeling, constantly adjusts insulin supply in real-time, thereby improving glucose level optimization in the body. This end-to-end method maximizes efficiency, providing personalized care and improved health outcomes, as confirmed by empirical evidence.
|
| 8 |
+
|
| 9 |
+
## Architecture
|
| 10 |
+
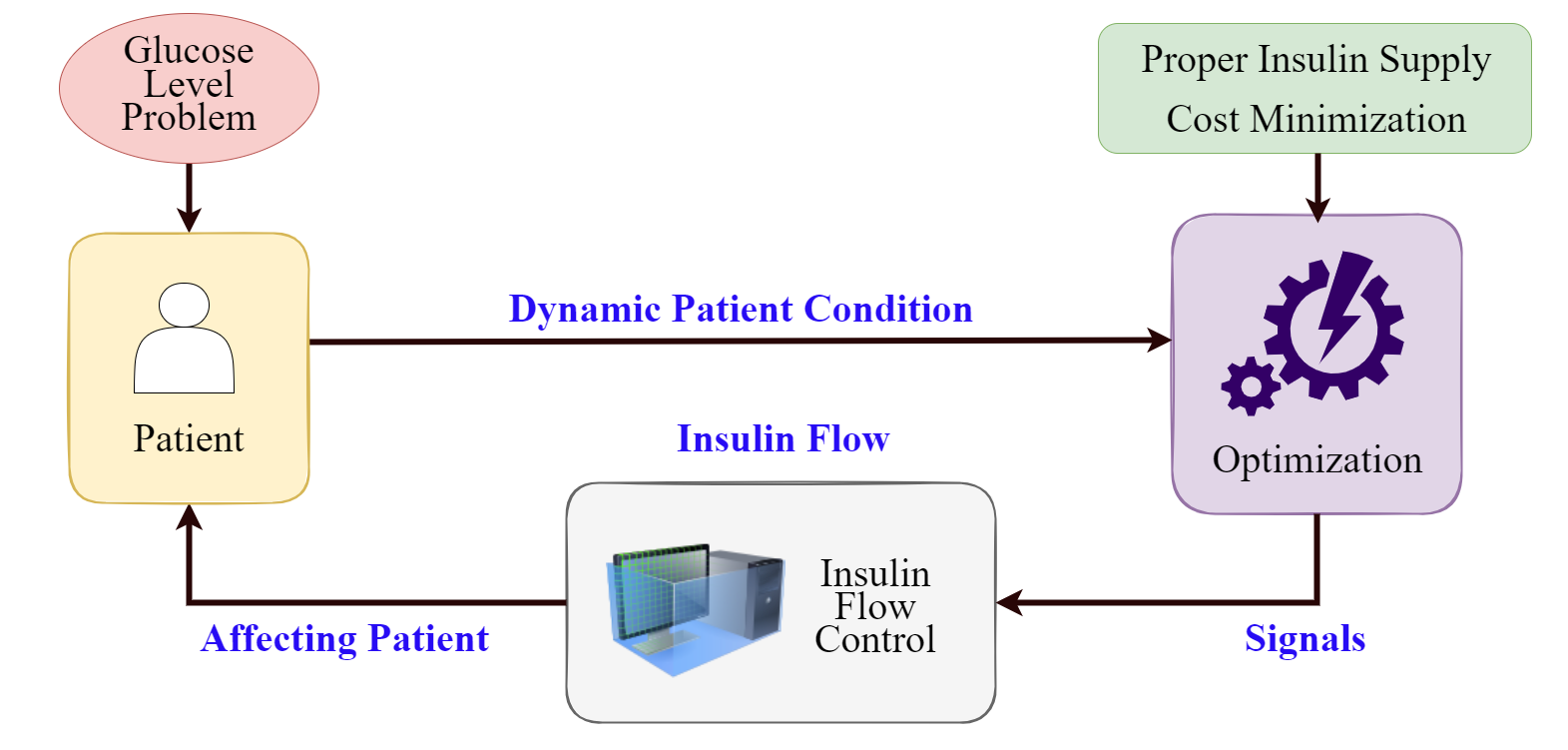
Pipeline of *NeuralCGMM*.
|
| 11 |
+
|
| 12 |
+
<p align="center">
|
| 13 |
+
<img src="fig/NeuralCGMM.png" width="1000"/>
|
| 14 |
+
</p>
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
<p align="center">
|
| 18 |
+
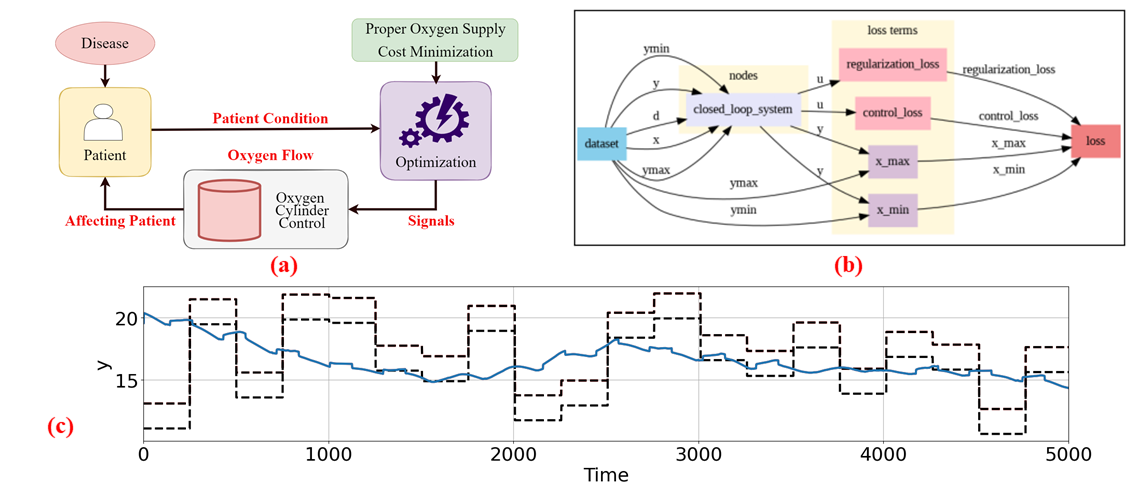
<img src="fig/allpic.png" width="1000"/>
|
| 19 |
+
</p>
|
| 20 |
+
|
| 21 |
+
|
| 22 |
+
---
|
| 23 |
+
|
| 24 |
+
## Citation
|
| 25 |
+
```
|
| 26 |
+
@inproceedings{
|
| 27 |
+
wasi2024neural,
|
| 28 |
+
title={Neural Control System for Continuous Glucose Monitoring and Maintenance},
|
| 29 |
+
author={Azmine Toushik Wasi},
|
| 30 |
+
booktitle={The Second Tiny Papers Track at ICLR 2024},
|
| 31 |
+
year={2024},
|
| 32 |
+
url={https://openreview.net/forum?id=Te4P3Cn54g}
|
| 33 |
+
}
|
| 34 |
+
```
|
fig/CLS.png
ADDED
|
fig/NeuralCGMM.png
ADDED
|
fig/allpic.png
ADDED
|
fig/output.png
ADDED
|
fig/problem.png
ADDED
|
model/README.md
ADDED
|
@@ -0,0 +1,9 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Neural Control System for Continuous Glucose Monitoring and Maintenance
|
| 2 |
+
|
| 3 |
+
The `main.py` has the full code for the simulation. Running this with all deendencies installed will train the model and run a synthetic simulation.
|
| 4 |
+
|
| 5 |
+
Synthetic test data and model is provided in `data` folder. By replacing them properly, the charts can be reproduced.
|
| 6 |
+
|
| 7 |
+
Also, a notebook is provided, too.
|
| 8 |
+
|
| 9 |
+
*We are very sorry for the unorganized code. Due to time constrains, we couldn't manage to organize fully.*
|
model/data/best_model.pt
ADDED
|
Binary file (12.8 kB). View file
|
|
|
model/data/test-data.pt
ADDED
|
Binary file (62.2 kB). View file
|
|
|
model/data/torch_dist.pt
ADDED
|
Binary file (37.2 kB). View file
|
|
|
model/dataloader.py
ADDED
|
@@ -0,0 +1,36 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import torch.nn as nn
|
| 3 |
+
from torch.utils.data import DataLoader
|
| 4 |
+
import numpy as np
|
| 5 |
+
|
| 6 |
+
import neuromancer.psl as psl
|
| 7 |
+
from neuromancer.system import Node, System
|
| 8 |
+
from neuromancer.modules import blocks
|
| 9 |
+
from neuromancer.dataset import DictDataset
|
| 10 |
+
from neuromancer.constraint import variable
|
| 11 |
+
from neuromancer.loss import PenaltyLoss
|
| 12 |
+
from neuromancer.problem import Problem
|
| 13 |
+
from neuromancer.trainer import Trainer
|
| 14 |
+
from neuromancer.plot import pltCL
|
| 15 |
+
|
| 16 |
+
def get_data(sys, nsteps, n_samples, xmin_range, batch_size, name="train"):
|
| 17 |
+
# sampled references for training the policy
|
| 18 |
+
batched_xmin = xmin_range.sample((n_samples, 1, nref)).repeat(1, nsteps + 1, 1)
|
| 19 |
+
batched_xmax = batched_xmin + 2.
|
| 20 |
+
|
| 21 |
+
# sampled disturbance trajectories from the simulation model
|
| 22 |
+
batched_dist = torch.stack([torch.tensor(sys.get_D(nsteps)) for _ in range(n_samples)])
|
| 23 |
+
|
| 24 |
+
# sampled initial conditions
|
| 25 |
+
batched_x0 = torch.stack([torch.tensor(sys.get_x0()).unsqueeze(0) for _ in range(n_samples)])
|
| 26 |
+
|
| 27 |
+
data = DictDataset(
|
| 28 |
+
{"x": batched_x0,
|
| 29 |
+
"y": batched_x0[:,:,[-1]],
|
| 30 |
+
"ymin": batched_xmin,
|
| 31 |
+
"ymax": batched_xmax,
|
| 32 |
+
"d": batched_dist},
|
| 33 |
+
name=name,
|
| 34 |
+
)
|
| 35 |
+
|
| 36 |
+
return DataLoader(data, batch_size=batch_size, collate_fn=data.collate_fn, shuffle=False)
|
model/main.py
ADDED
|
@@ -0,0 +1,176 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import torch.nn as nn
|
| 3 |
+
from torch.utils.data import DataLoader
|
| 4 |
+
import numpy as np
|
| 5 |
+
from math import *
|
| 6 |
+
|
| 7 |
+
import neuromancer.psl as psl
|
| 8 |
+
from neuromancer.system import Node, System
|
| 9 |
+
from neuromancer.modules import blocks
|
| 10 |
+
from neuromancer.dataset import DictDataset
|
| 11 |
+
from neuromancer.constraint import variable
|
| 12 |
+
from neuromancer.loss import PenaltyLoss
|
| 13 |
+
from neuromancer.problem import Problem
|
| 14 |
+
from neuromancer.trainer import Trainer
|
| 15 |
+
from neuromancer.plot import pltCL
|
| 16 |
+
|
| 17 |
+
from dataloader import get_data
|
| 18 |
+
|
| 19 |
+
|
| 20 |
+
|
| 21 |
+
# ground truth system model
|
| 22 |
+
sys = psl.systems['LinearSimpleSingleZone']()
|
| 23 |
+
|
| 24 |
+
# problem dimensions
|
| 25 |
+
nx = sys.nx # number of states
|
| 26 |
+
nu = sys.nu # number of control inputs
|
| 27 |
+
nd = sys.nD # number of disturbances
|
| 28 |
+
nd_obs = sys.nD_obs # number of observable disturbances
|
| 29 |
+
ny = sys.ny # number of controlled outputs
|
| 30 |
+
nref = ny # number of references
|
| 31 |
+
|
| 32 |
+
# control action bounds
|
| 33 |
+
umin = torch.tensor(sys.umin)
|
| 34 |
+
umax = torch.tensor(sys.umax)
|
| 35 |
+
|
| 36 |
+
# Data
|
| 37 |
+
nsteps = 100 # prediction horizon
|
| 38 |
+
n_samples = 2000 # number of sampled scenarios
|
| 39 |
+
batch_size = 64
|
| 40 |
+
|
| 41 |
+
# range for lower comfort bound
|
| 42 |
+
xmin_range = torch.distributions.Uniform(18., 20.)
|
| 43 |
+
|
| 44 |
+
train_loader, dev_loader = [
|
| 45 |
+
get_data(sys, nsteps, n_samples, xmin_range, batch_size, name=name)
|
| 46 |
+
for name in ("train", "dev")
|
| 47 |
+
]
|
| 48 |
+
|
| 49 |
+
# SSM
|
| 50 |
+
# extract exact state space model matrices:
|
| 51 |
+
A = torch.tensor(sys.A)
|
| 52 |
+
B = torch.tensor(sys.Beta)
|
| 53 |
+
C = torch.tensor(sys.C)
|
| 54 |
+
E = torch.tensor(sys.E)
|
| 55 |
+
|
| 56 |
+
|
| 57 |
+
# state-space model of the building dynamics:
|
| 58 |
+
# x_k+1 = A x_k + B u_k + E d_k
|
| 59 |
+
xnext = lambda x, u, d: x @ A.T + u @ B.T + d @ E.T
|
| 60 |
+
state_model = Node(xnext, ['x', 'u', 'd'], ['x'], name='SSM')
|
| 61 |
+
|
| 62 |
+
# y_k = C x_k
|
| 63 |
+
ynext = lambda x: x @ C.T
|
| 64 |
+
output_model = Node(ynext, ['x'], ['y'], name='y=Cx')
|
| 65 |
+
|
| 66 |
+
# partially observable disturbance model
|
| 67 |
+
dist_model = lambda d: d[:, sys.d_idx]
|
| 68 |
+
patient_cond_change = Node(dist_model, ['d'], ['patient_obs'], name='patient_cond_change')
|
| 69 |
+
|
| 70 |
+
# neural net control policy
|
| 71 |
+
net = blocks.MLP_bounds(
|
| 72 |
+
insize=ny + 2*nref + nd_obs,
|
| 73 |
+
outsize=nu,
|
| 74 |
+
hsizes=[32, 32],
|
| 75 |
+
nonlin=nn.GELU,
|
| 76 |
+
min=umin,
|
| 77 |
+
max=umax,
|
| 78 |
+
)
|
| 79 |
+
policy = Node(net, ['y', 'ymin', 'ymax', 'patient_obs'], ['u'], name='policy')
|
| 80 |
+
|
| 81 |
+
# closed-loop system model
|
| 82 |
+
closed_loop_system = System([patient_cond_change, policy, state_model, output_model],
|
| 83 |
+
nsteps=nsteps,
|
| 84 |
+
name='closed_loop_system')
|
| 85 |
+
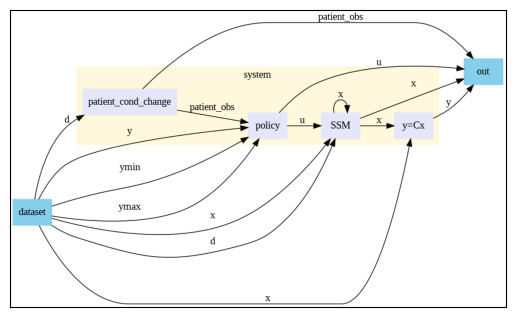
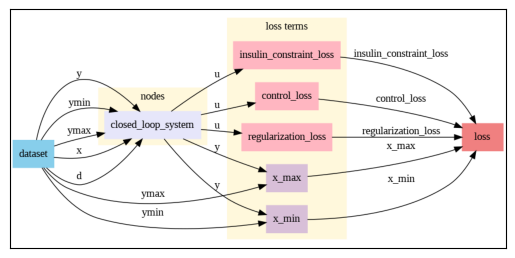
closed_loop_system.show()
|
| 86 |
+
|
| 87 |
+
# dpc
|
| 88 |
+
# variables
|
| 89 |
+
y = variable('y')
|
| 90 |
+
u = variable('u')
|
| 91 |
+
ymin = variable('ymin')
|
| 92 |
+
ymax = variable('ymax')
|
| 93 |
+
|
| 94 |
+
# objectives
|
| 95 |
+
action_loss = 0.01 * (u == 0.0) # energy minimization
|
| 96 |
+
du_loss = 0.1 * (u[:,:-1,:] - u[:,1:,:] == 0.0) # delta u minimization to prevent agressive changes in control actions
|
| 97 |
+
action_limit_loss = 0.02 * (abs(u[:, 1:, :] - u[:, :-1, :])==0.0) # constraint loss for insulin delivery
|
| 98 |
+
|
| 99 |
+
# thermal comfort constraints
|
| 100 |
+
state_lower_bound_penalty = 50.*(y > ymin)
|
| 101 |
+
state_upper_bound_penalty = 50.*(y < ymax)
|
| 102 |
+
|
| 103 |
+
# objectives and constraints names for nicer plot
|
| 104 |
+
action_loss.name = 'control_loss'
|
| 105 |
+
du_loss.name = 'regularization_loss'
|
| 106 |
+
action_limit_loss.name = 'insulin_constraint_loss'
|
| 107 |
+
state_lower_bound_penalty.name = 'x_min'
|
| 108 |
+
state_upper_bound_penalty.name = 'x_max'
|
| 109 |
+
|
| 110 |
+
# list of constraints and objectives
|
| 111 |
+
objectives = [action_loss, du_loss, action_limit_loss]
|
| 112 |
+
constraints = [state_lower_bound_penalty, state_upper_bound_penalty]
|
| 113 |
+
|
| 114 |
+
# data (x_k, r_k) -> parameters (xi_k) -> policy (u_k) -> dynamics (x_k+1)
|
| 115 |
+
nodes = [closed_loop_system]
|
| 116 |
+
# create constrained optimization loss
|
| 117 |
+
loss = PenaltyLoss(objectives, constraints)
|
| 118 |
+
# construct constrained optimization problem
|
| 119 |
+
problem = Problem(nodes, loss)
|
| 120 |
+
# plot computational graph
|
| 121 |
+
problem.show()
|
| 122 |
+
|
| 123 |
+
|
| 124 |
+
optimizer = torch.optim.AdamW(problem.parameters(), lr=0.001)
|
| 125 |
+
# Neuromancer trainer
|
| 126 |
+
trainer = Trainer(
|
| 127 |
+
problem,
|
| 128 |
+
train_loader,
|
| 129 |
+
dev_loader,
|
| 130 |
+
optimizer=optimizer,
|
| 131 |
+
epochs=200,
|
| 132 |
+
train_metric='train_loss',
|
| 133 |
+
eval_metric='dev_loss',
|
| 134 |
+
warmup=50,
|
| 135 |
+
)
|
| 136 |
+
|
| 137 |
+
|
| 138 |
+
# Train control policy
|
| 139 |
+
best_model = trainer.train()
|
| 140 |
+
# load best trained model
|
| 141 |
+
trainer.model.load_state_dict(best_model)
|
| 142 |
+
|
| 143 |
+
|
| 144 |
+
#RANDOM TEST
|
| 145 |
+
|
| 146 |
+
nsteps_test = 3000
|
| 147 |
+
|
| 148 |
+
# generate reference
|
| 149 |
+
np_refs = psl.signals.step(nsteps_test+1, 1, min=18, max=21, randsteps=8)
|
| 150 |
+
ymin_val = torch.tensor(np_refs, dtype=torch.float32).reshape(1, nsteps_test+1, 1)
|
| 151 |
+
ymax_val = ymin_val+6.0
|
| 152 |
+
# generate disturbance signal
|
| 153 |
+
torch_dist = torch.tensor(sys.get_D(nsteps_test+1)).unsqueeze(0)
|
| 154 |
+
# initial data for closed loop simulation
|
| 155 |
+
x0 = torch.tensor(sys.get_x0()).reshape(1, 1, nx)
|
| 156 |
+
data = {'x': x0,
|
| 157 |
+
'y': x0[:, :, [-1]],
|
| 158 |
+
'ymin': ymin_val,
|
| 159 |
+
'ymax': ymax_val,
|
| 160 |
+
'd': torch_dist}
|
| 161 |
+
closed_loop_system.nsteps = nsteps_test
|
| 162 |
+
# perform closed-loop simulation
|
| 163 |
+
trajectories = closed_loop_system(data)
|
| 164 |
+
|
| 165 |
+
# constraints bounds
|
| 166 |
+
Umin = umin * np.ones([nsteps_test, nu])
|
| 167 |
+
Umax = umax * np.ones([nsteps_test, nu])
|
| 168 |
+
Ymin = trajectories['ymin'].detach().reshape(nsteps_test+1, nref)
|
| 169 |
+
Ymax = trajectories['ymax'].detach().reshape(nsteps_test+1, nref)
|
| 170 |
+
# plot closed loop trajectories
|
| 171 |
+
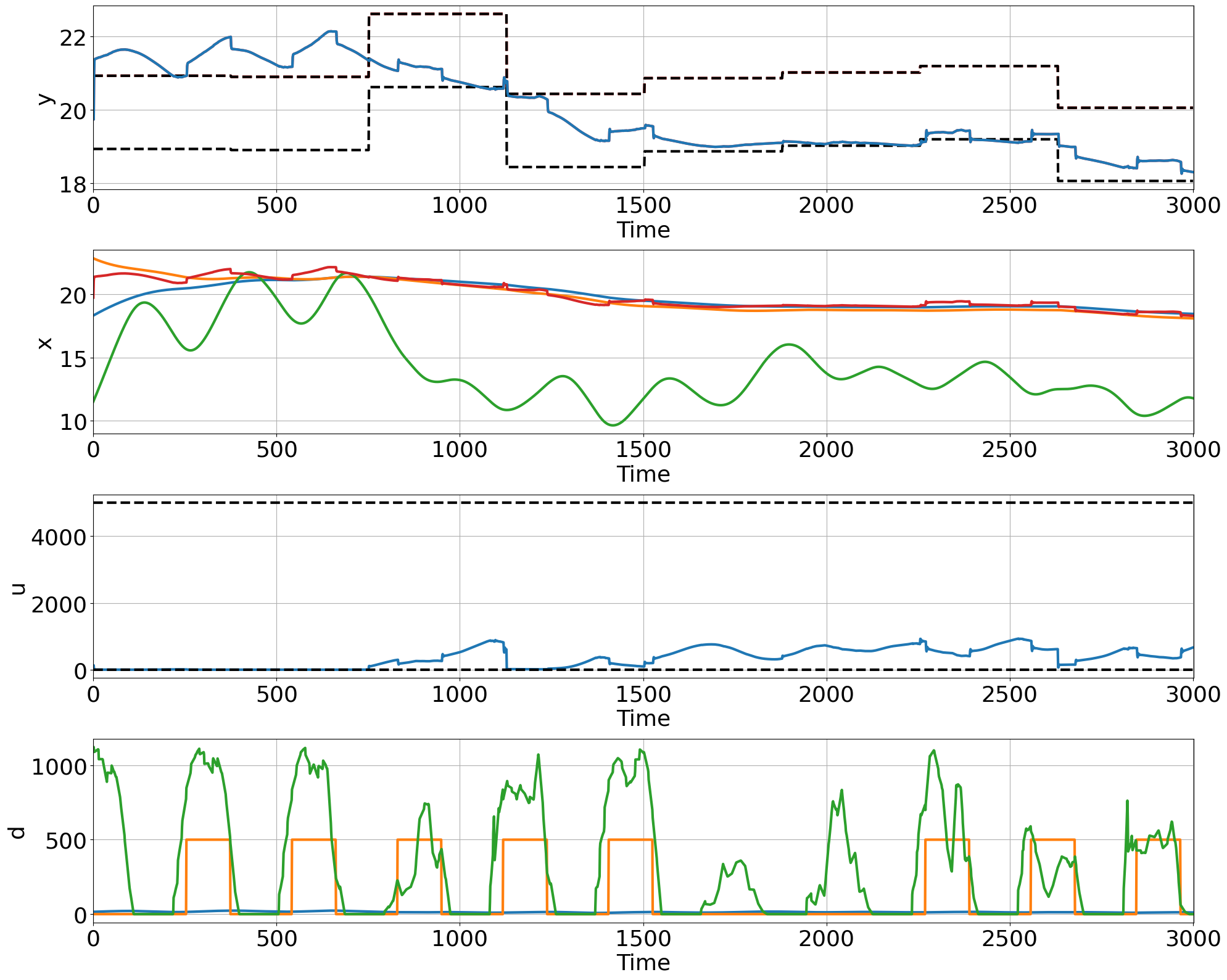
pltCL(Y=trajectories['y'].detach().reshape(nsteps_test+1, ny),
|
| 172 |
+
R=Ymax,
|
| 173 |
+
X=trajectories['x'].detach().reshape(nsteps_test+1, nx),
|
| 174 |
+
D=trajectories['d'].detach().reshape(nsteps_test+1, nd),
|
| 175 |
+
U=trajectories['u'].detach().reshape(nsteps_test, nu),
|
| 176 |
+
Umin=Umin, Umax=Umax, Ymin=Ymin, Ymax=Ymax)
|
model/models.md
ADDED
|
@@ -0,0 +1,12 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Neuromancer Install
|
| 2 |
+
### Install (Colab only)
|
| 3 |
+
# Skip this step when running locally.
|
| 4 |
+
|
| 5 |
+
```
|
| 6 |
+
pip install "neuromancer[examples] @ git+https://github.com/pnnl/neuromancer.git@master"
|
| 7 |
+
```
|
| 8 |
+
|
| 9 |
+
# General Method
|
| 10 |
+
```
|
| 11 |
+
pip install neurormancer
|
| 12 |
+
```
|