update
Browse files- README.md +6 -4
- extras/tissue_slice_frontal.png +0 -0
- requirements.txt +2 -1
- vis_hit_sample.py +82 -1
README.md
CHANGED
|
@@ -70,16 +70,18 @@ print(next(iter(male_train)))
|
|
| 70 |
```
|
| 71 |
|
| 72 |
### Visualize data
|
|
|
|
| 73 |
```angular2html
|
| 74 |
-
pip install
|
| 75 |
```
|
| 76 |
```angular2html
|
| 77 |
-
|
| 78 |
```
|
|
|
|
| 79 |
```angular2html
|
| 80 |
-
python vis_hit_sample.py --gender male --split test --idx 5
|
| 81 |
```
|
| 82 |
-

|
| 81 |
```angular2html
|
| 82 |
+
python vis_hit_sample.py --gender male --split test --idx 5 --show_tissue
|
| 83 |
```
|
| 84 |
+

|
| 85 |
|
| 86 |
## Dataset Structure
|
| 87 |
The dataset is structured as follows:
|
extras/tissue_slice_frontal.png
ADDED
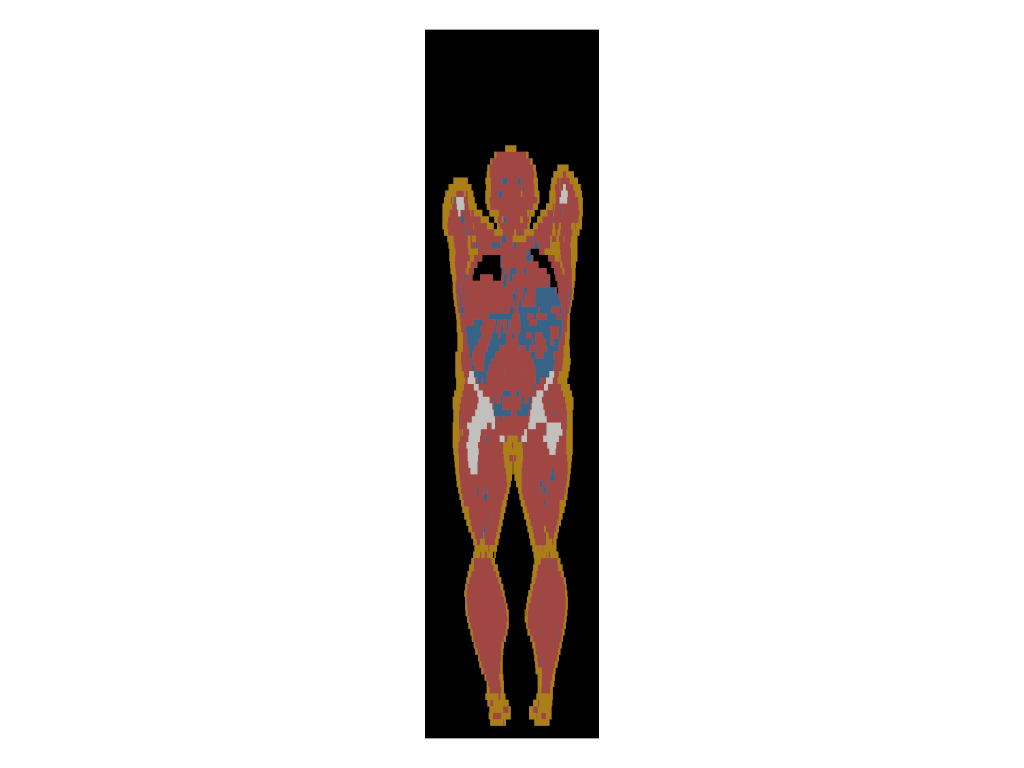
|
requirements.txt
CHANGED
|
@@ -1,2 +1,3 @@
|
|
| 1 |
datasets
|
| 2 |
-
open3d
|
|
|
|
|
|
| 1 |
datasets
|
| 2 |
+
open3d
|
| 3 |
+
pyvista
|
vis_hit_sample.py
CHANGED
|
@@ -3,10 +3,77 @@ import argparse
|
|
| 3 |
from datasets import load_dataset
|
| 4 |
|
| 5 |
import open3d as o3d
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 6 |
import numpy as np
|
| 7 |
import random
|
| 8 |
|
| 9 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 10 |
def vis_hit_sample(sample):
|
| 11 |
"""
|
| 12 |
:param sample: HIT dataset sample
|
|
@@ -65,6 +132,8 @@ if __name__ == '__main__':
|
|
| 65 |
parser.add_argument('--gender', type=str, default='male')
|
| 66 |
parser.add_argument('--split', type=str, default='train')
|
| 67 |
parser.add_argument('--idx', type=int, default=None)
|
|
|
|
|
|
|
| 68 |
|
| 69 |
# get args
|
| 70 |
args = parser.parse_args()
|
|
@@ -98,4 +167,16 @@ if __name__ == '__main__':
|
|
| 98 |
hit_sample = hit_dataset[idx]
|
| 99 |
# visualize the sample
|
| 100 |
print(f"Visualizing sample no. {idx} in {args.gender}:{args.split}.")
|
| 101 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 3 |
from datasets import load_dataset
|
| 4 |
|
| 5 |
import open3d as o3d
|
| 6 |
+
import pyvista as pv
|
| 7 |
+
from PIL import Image
|
| 8 |
+
import matplotlib
|
| 9 |
+
import matplotlib.pyplot as plt
|
| 10 |
+
|
| 11 |
import numpy as np
|
| 12 |
import random
|
| 13 |
|
| 14 |
|
| 15 |
+
def plot_3D_image(values, resolution, p=None, interactive_slice=False, orthogonal_slices=True):
|
| 16 |
+
''' Interactive plot of the 3D volume'''
|
| 17 |
+
|
| 18 |
+
# Create the spatial reference
|
| 19 |
+
grid = pv.ImageData()
|
| 20 |
+
values = np.transpose(values, (1,2,0))
|
| 21 |
+
# Set the grid dimensions: shape + 1 because we want to inject our values on
|
| 22 |
+
# the CELL data
|
| 23 |
+
grid.dimensions = np.array(values.shape) + 1
|
| 24 |
+
|
| 25 |
+
# Edit the spatial reference
|
| 26 |
+
# The bottom left corner of the data set
|
| 27 |
+
origin = np.array(resolution[0]) * np.array(values.shape) * 0.5
|
| 28 |
+
grid.origin = origin
|
| 29 |
+
#print(f'Scan size in meter: {origin * 2}')
|
| 30 |
+
grid.spacing = resolution[0] # These are the cell sizes along each axis
|
| 31 |
+
|
| 32 |
+
# Add the data values to the cell data
|
| 33 |
+
grid.cell_data["values"] = values.flatten(order="F") # Flatten the array!
|
| 34 |
+
|
| 35 |
+
if p is None:
|
| 36 |
+
p = pv.Plotter()
|
| 37 |
+
|
| 38 |
+
if orthogonal_slices:
|
| 39 |
+
slices = grid.slice_orthogonal()
|
| 40 |
+
cmap = matplotlib.colors.ListedColormap(['black', 'indianred', 'goldenrod', 'steelblue', 'ghostwhite'])
|
| 41 |
+
p.add_mesh(slices, cmap=cmap)
|
| 42 |
+
|
| 43 |
+
if interactive_slice:
|
| 44 |
+
p.add_mesh_clip_plane(grid)
|
| 45 |
+
|
| 46 |
+
return p
|
| 47 |
+
|
| 48 |
+
|
| 49 |
+
def get_sliced_mri_png(sample):
|
| 50 |
+
# get data
|
| 51 |
+
mri = np.asarray(sample['mri_seg'])
|
| 52 |
+
resolution = np.asarray(sample['resolution'])
|
| 53 |
+
# set plotter
|
| 54 |
+
p = pv.Plotter(shape=(1, 1), off_screen=True)
|
| 55 |
+
p.subplot(0, 0)
|
| 56 |
+
plotter = plot_3D_image(mri, resolution, p, interactive_slice=False, orthogonal_slices=True)
|
| 57 |
+
plotter.view_yz()
|
| 58 |
+
plotter.remove_scalar_bar()
|
| 59 |
+
# store screenshot
|
| 60 |
+
img = p.screenshot("./extras/img.png", return_img=True)
|
| 61 |
+
# read screenshot
|
| 62 |
+
img = Image.fromarray(img)
|
| 63 |
+
# set plotter lateral
|
| 64 |
+
p = pv.Plotter(shape=(1, 1), off_screen=True)
|
| 65 |
+
p.subplot(0, 0)
|
| 66 |
+
plotter = plot_3D_image(mri, resolution, p, interactive_slice=False, orthogonal_slices=True)
|
| 67 |
+
plotter.remove_scalar_bar()
|
| 68 |
+
plotter.view_xz()
|
| 69 |
+
img_lateral = p.screenshot("./extras/img_lateral.png", return_img=True)
|
| 70 |
+
img_lateral = Image.fromarray(img_lateral)
|
| 71 |
+
# resize
|
| 72 |
+
img = img.resize((512+128, 372+128))
|
| 73 |
+
img_lateral = img_lateral.resize((512+128, 372+128))
|
| 74 |
+
return img, img_lateral
|
| 75 |
+
|
| 76 |
+
|
| 77 |
def vis_hit_sample(sample):
|
| 78 |
"""
|
| 79 |
:param sample: HIT dataset sample
|
|
|
|
| 132 |
parser.add_argument('--gender', type=str, default='male')
|
| 133 |
parser.add_argument('--split', type=str, default='train')
|
| 134 |
parser.add_argument('--idx', type=int, default=None)
|
| 135 |
+
parser.add_argument('--show_skin', action='store_true')
|
| 136 |
+
parser.add_argument('--show_tissue', action='store_true')
|
| 137 |
|
| 138 |
# get args
|
| 139 |
args = parser.parse_args()
|
|
|
|
| 167 |
hit_sample = hit_dataset[idx]
|
| 168 |
# visualize the sample
|
| 169 |
print(f"Visualizing sample no. {idx} in {args.gender}:{args.split}.")
|
| 170 |
+
if args.show_tissue:
|
| 171 |
+
img, img_lateral = get_sliced_mri_png(hit_sample)
|
| 172 |
+
img.show()
|
| 173 |
+
img_lateral.show()
|
| 174 |
+
elif args.show_skin:
|
| 175 |
+
vis_hit_sample(hit_sample)
|
| 176 |
+
else:
|
| 177 |
+
img, img_lateral = get_sliced_mri_png(hit_sample)
|
| 178 |
+
img.show()
|
| 179 |
+
img_lateral.show()
|
| 180 |
+
vis_hit_sample(hit_sample)
|
| 181 |
+
|
| 182 |
+
|