File size: 7,612 Bytes
139850b b243c9e 139850b dd88eb7 ef9db36 139850b b243c9e 139850b dd88eb7 139850b dd88eb7 139850b dd88eb7 139850b dd88eb7 139850b dd88eb7 139850b 9b1d3a3 dd88eb7 ad30235 fb61f80 ad30235 fb61f80 ad30235 dd88eb7 139850b dd88eb7 139850b dd88eb7 9b1d3a3 139850b 0238d7d 139850b 0e72621 dd88eb7 9b1d3a3 139850b 0238d7d 139850b dd88eb7 139850b dd88eb7 9b1d3a3 139850b 0238d7d 139850b dd88eb7 9b1d3a3 139850b 0238d7d 139850b ad30235 139850b 32f3b11 |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 |
# Model Overview
A pre-trained model for volumetric (3D) segmentation of brain tumor subregions from multimodal MRIs based on BraTS 2018 data.
The model is trained to segment 3 nested subregions of primary brain tumors (gliomas): the "enhancing tumor" (ET), the "tumor core" (TC), the "whole tumor" (WT) based on 4 aligned input MRI scans (T1c, T1, T2, FLAIR).
- The ET is described by areas that show hyper intensity in T1c when compared to T1, but also when compared to "healthy" white matter in T1c.
- The TC describes the bulk of the tumor, which is what is typically resected. The TC entails the ET, as well as the necrotic (fluid-filled) and the non-enhancing (solid) parts of the tumor.
- The WT describes the complete extent of the disease, as it entails the TC and the peritumoral edema (ED), which is typically depicted by hyper-intense signal in FLAIR.

## Data
The training data is from the [Multimodal Brain Tumor Segmentation Challenge (BraTS) 2018](https://www.med.upenn.edu/sbia/brats2018.html).
- Target: 3 tumor subregions
- Task: Segmentation
- Modality: MRI
- Size: 285 3D volumes (4 channels each)
The provided labelled data was partitioned, based on our own split, into training (200 studies), validation (42 studies) and testing (43 studies) datasets.
### Preprocessing
The data list/split can be created with the script `scripts/prepare_datalist.py`.
```
python scripts/prepare_datalist.py --path your-brats18-dataset-path
```
## Training configuration
This model utilized a similar approach described in 3D MRI brain tumor segmentation using autoencoder regularization, which was a winning method in BraTS2018 [1]. The training was performed with the following:
- GPU: At least 16GB of GPU memory.
- Actual Model Input: 224 x 224 x 144
- AMP: True
- Optimizer: Adam
- Learning Rate: 1e-4
- Loss: DiceLoss
## Input
4 channel aligned MRIs at 1 x 1 x 1 mm
- T1c
- T1
- T2
- FLAIR
## Output
3 channels
- Label 0: TC tumor subregion
- Label 1: WT tumor subregion
- Label 2: ET tumor subregion
## Performance
Dice score was used for evaluating the performance of the model. This model achieved Dice scores on the validation data of:
- Tumor core (TC): 0.8559
- Whole tumor (WT): 0.9026
- Enhancing tumor (ET): 0.7905
- Average: 0.8518
Please note that this bundle is non-deterministic because of the trilinear interpolation used in the network. Therefore, reproducing the training process may not get exactly the same performance.
Please refer to https://pytorch.org/docs/stable/notes/randomness.html#reproducibility for more details about reproducibility.
#### Training Loss and Dice
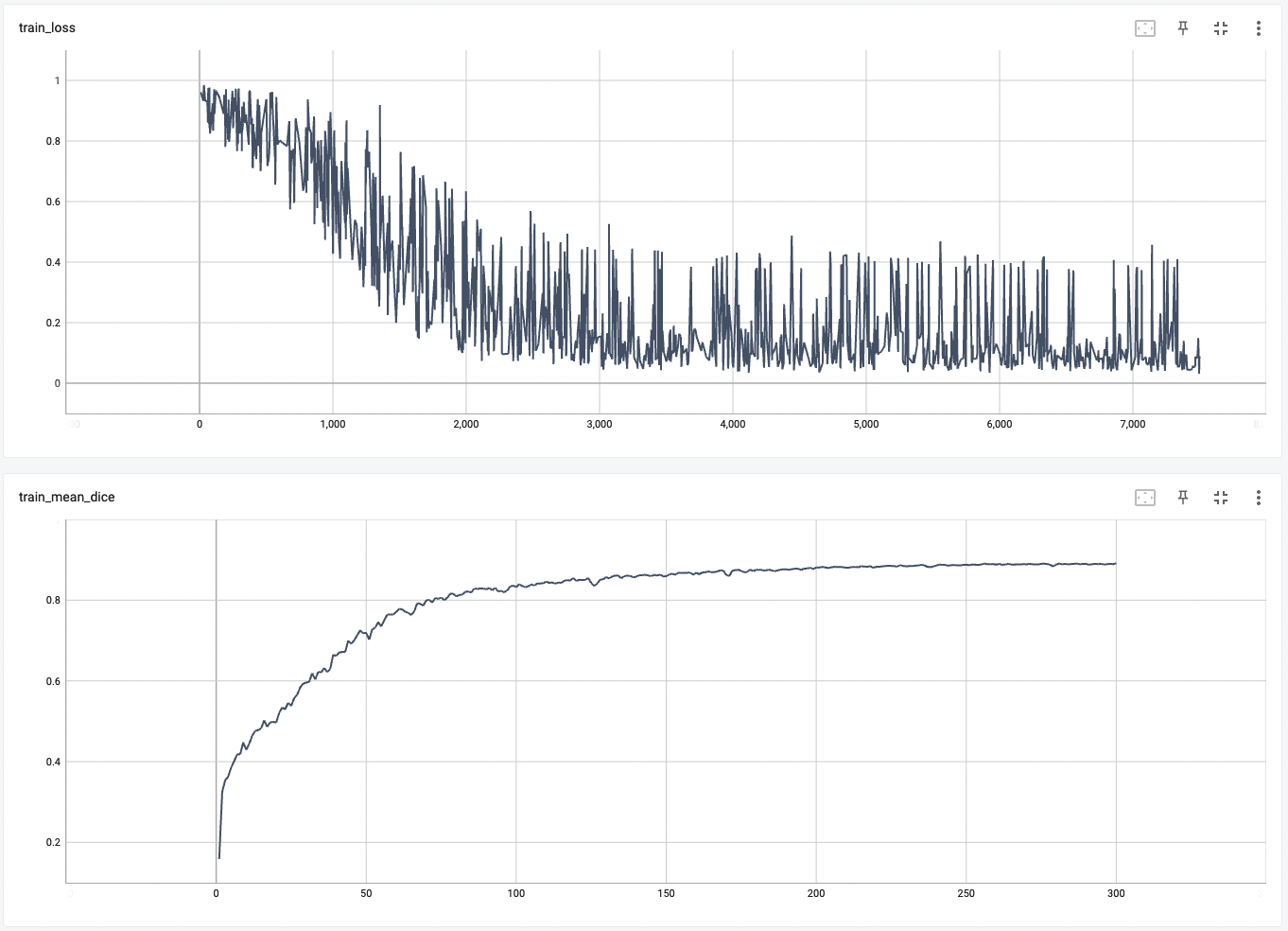
#### Validation Dice
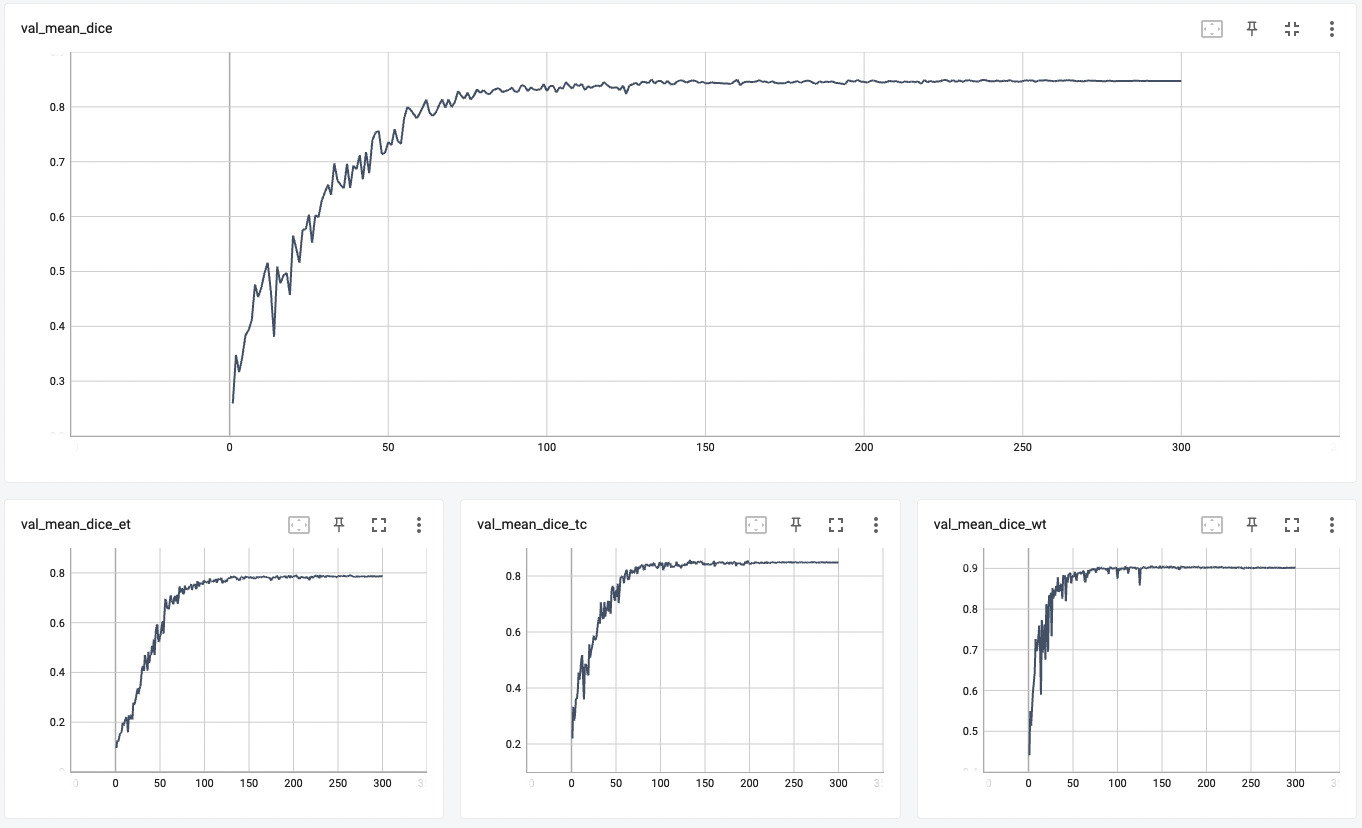
#### TensorRT speedup
The `brats_mri_segmentation` bundle supports acceleration with TensorRT through the ONNX-TensorRT method. The table below displays the speedup ratios observed on an A100 80G GPU.
| method | torch_fp32(ms) | torch_amp(ms) | trt_fp32(ms) | trt_fp16(ms) | speedup amp | speedup fp32 | speedup fp16 | amp vs fp16|
| :---: | :---: | :---: | :---: | :---: | :---: | :---: | :---: | :---: |
| model computation | 5.49 | 4.36 | 2.35 | 2.09 | 1.26 | 2.34 | 2.63 | 2.09 |
| end2end | 592.01 | 434.59 | 395.73 | 394.93 | 1.36 | 1.50 | 1.50 | 1.10 |
Where:
- `model computation` means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
- `end2end` means run the bundle end-to-end with the TensorRT based model.
- `torch_fp32` and `torch_amp` are for the PyTorch models with or without `amp` mode.
- `trt_fp32` and `trt_fp16` are for the TensorRT based models converted in corresponding precision.
- `speedup amp`, `speedup fp32` and `speedup fp16` are the speedup ratios of corresponding models versus the PyTorch float32 model
- `amp vs fp16` is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
Currently, the only available method to accelerate this model is through ONNX-TensorRT. However, the Torch-TensorRT method is under development and will be available in the near future.
This result is benchmarked under:
- TensorRT: 8.5.3+cuda11.8
- Torch-TensorRT Version: 1.4.0
- CPU Architecture: x86-64
- OS: ubuntu 20.04
- Python version:3.8.10
- CUDA version: 12.0
- GPU models and configuration: A100 80G
## MONAI Bundle Commands
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
#### Execute training:
```
python -m monai.bundle run --config_file configs/train.json
```
Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using `--dataset_dir`:
```
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path>
```
#### Override the `train` config to execute multi-GPU training:
```
torchrun --standalone --nnodes=1 --nproc_per_node=8 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']"
```
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove `--standalone`, modify `--nnodes`, or do some other necessary changes according to the machine used. For more details, please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html).
#### Override the `train` config to execute evaluation with the trained model:
```
python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']"
```
#### Execute inference:
```
python -m monai.bundle run --config_file configs/inference.json
```
#### Export checkpoint to TensorRT based models with fp32 or fp16 precision:
```bash
python -m monai.bundle trt_export --net_id network_def \
--filepath models/model_trt.ts --ckpt_file models/model.pt \
--meta_file configs/metadata.json --config_file configs/inference.json \
--precision <fp32/fp16> --input_shape "[1, 4, 240, 240, 160]" --use_onnx "True" \
--use_trace "True"
```
#### Execute inference with the TensorRT model:
```
python -m monai.bundle run --config_file "['configs/inference.json', 'configs/inference_trt.json']"
```
# References
[1] Myronenko, Andriy. "3D MRI brain tumor segmentation using autoencoder regularization." International MICCAI Brainlesion Workshop. Springer, Cham, 2018. https://arxiv.org/abs/1810.11654.
# License
Copyright (c) MONAI Consortium
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
|