retrain using an internal pretrained ResNet18
Browse files- README.md +22 -2
- configs/inference.json +0 -3
- configs/metadata.json +5 -3
- configs/multi_gpu_train.json +1 -2
- configs/train.json +1 -2
- docs/README.md +22 -2
- models/model.pt +1 -1
README.md
CHANGED
|
@@ -46,6 +46,21 @@ The training was performed with the following:
|
|
| 46 |
- Loss: BCEWithLogitsLoss
|
| 47 |
- Whole slide image reader: cuCIM (if running on Windows or Mac, please install `OpenSlide` on your system and change `wsi_reader` to "OpenSlide")
|
| 48 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 49 |
### Input
|
| 50 |
|
| 51 |
The training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
|
|
@@ -58,12 +73,16 @@ A probability number of the input patch being tumor or normal.
|
|
| 58 |
|
| 59 |
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
|
| 60 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 61 |
## Performance
|
| 62 |
|
| 63 |
FROC score is used for evaluating the performance of the model. After inference is done, `evaluate_froc.sh` needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
|
| 64 |
-
|
| 65 |
|
| 66 |
-

|
| 48 |
|
| 49 |
+
### Pretrained Weights
|
| 50 |
+
|
| 51 |
+
By setting the `"pretrained"` parameter of `TorchVisionFCModel` in the config file to `true`, ImageNet pre-trained weights will be used for training. Please note that these weights are for non-commercial use. Each user is responsible for checking the content of the models/datasets and the applicable licenses and determining if suitable for the intended use. In order to use other pretrained weights, you can use `CheckpointLoader` in train handlers section as the first handler:
|
| 52 |
+
|
| 53 |
+
```json
|
| 54 |
+
{
|
| 55 |
+
"_target_": "CheckpointLoader",
|
| 56 |
+
"load_path": "$@bundle_root + '/pretrained_resnet18.pth'",
|
| 57 |
+
"strict": false,
|
| 58 |
+
"load_dict": {
|
| 59 |
+
"model_new": "@network"
|
| 60 |
+
}
|
| 61 |
+
}
|
| 62 |
+
```
|
| 63 |
+
|
| 64 |
### Input
|
| 65 |
|
| 66 |
The training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
|
|
|
|
| 73 |
|
| 74 |
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
|
| 75 |
|
| 76 |
+
### Note on determinism
|
| 77 |
+
|
| 78 |
+
By default this bundle use a deterministic approach to make the results reproducible. However, it comes at a cost of performance loss. Thus if you do not care about reproducibility, you can have a performance gain by replacing `"$monai.utils.set_determinism"` line with `"$setattr(torch.backends.cudnn, 'benchmark', True)"` in initialize section of training configuration (`configs/train.json` and `configs/multi_gpu_train.json` for single GPU and multi-GPU training respectively).
|
| 79 |
+
|
| 80 |
## Performance
|
| 81 |
|
| 82 |
FROC score is used for evaluating the performance of the model. After inference is done, `evaluate_froc.sh` needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
|
| 83 |
+
Using an internal pretrained weights for ResNet18, this model deterministically achieves the 0.90 accuracy on validation patches, and FROC of 0.72 on the 48 Camelyon testing data that have ground truth annotations available.
|
| 84 |
|
| 85 |
+
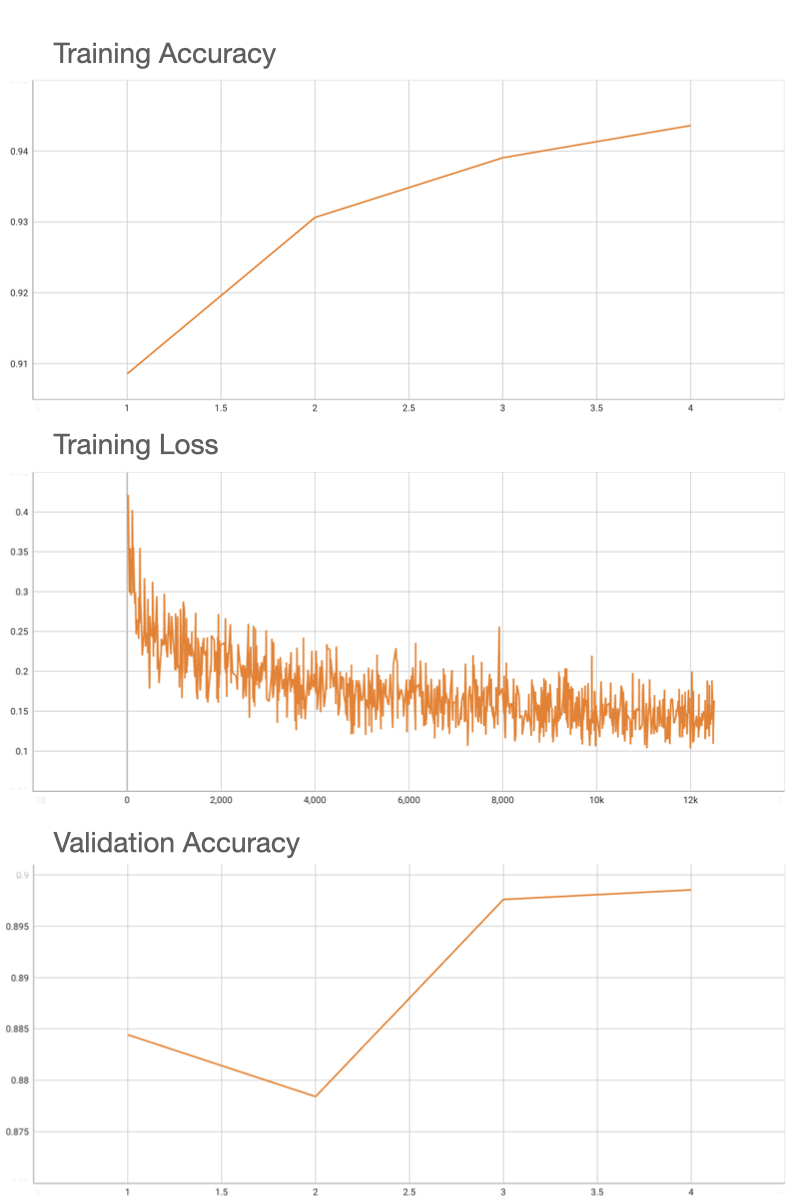
|
| 86 |
|
| 87 |
The `pathology_tumor_detection` bundle supports acceleration with TensorRT. The table below displays the speedup ratios observed on an A100 80G GPU.
|
| 88 |
|
|
|
|
| 94 |
| end2end |224.97 | 223.50 | 222.65 | 224.03 | 1.01 | 1.01 | 1.00 | 1.00 |
|
| 95 |
|
| 96 |
Where:
|
| 97 |
+
|
| 98 |
- `model computation` means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
|
| 99 |
- `end2end` means run the bundle end-to-end with the TensorRT based model.
|
| 100 |
- `torch_fp32` and `torch_amp` are for the PyTorch models with or without `amp` mode.
|
configs/inference.json
CHANGED
|
@@ -125,9 +125,6 @@
|
|
| 125 |
"amp": true,
|
| 126 |
"decollate": false
|
| 127 |
},
|
| 128 |
-
"initialize": [
|
| 129 |
-
"$setattr(torch.backends.cudnn, 'benchmark', True)"
|
| 130 |
-
],
|
| 131 |
"run": [
|
| 132 |
"$@evaluator.run()"
|
| 133 |
]
|
|
|
|
| 125 |
"amp": true,
|
| 126 |
"decollate": false
|
| 127 |
},
|
|
|
|
|
|
|
|
|
|
| 128 |
"run": [
|
| 129 |
"$@evaluator.run()"
|
| 130 |
]
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,9 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.5.
|
| 4 |
"changelog": {
|
|
|
|
|
|
|
| 5 |
"0.5.2": "update TensorRT descriptions",
|
| 6 |
"0.5.1": "update the TensorRT part in the README file",
|
| 7 |
"0.5.0": "add the command of executing inference with TensorRT models",
|
|
@@ -42,8 +44,8 @@
|
|
| 42 |
"label_classes": "binary labels for each patch",
|
| 43 |
"pred_classes": "scalar probability",
|
| 44 |
"eval_metrics": {
|
| 45 |
-
"accuracy": 0.
|
| 46 |
-
"froc": 0.
|
| 47 |
},
|
| 48 |
"intended_use": "This is an example, not to be used for diagnostic purposes",
|
| 49 |
"references": [
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.5.4",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.5.4": "retrain using an internal pretrained ResNet18",
|
| 6 |
+
"0.5.3": "make the training bundle deterministic",
|
| 7 |
"0.5.2": "update TensorRT descriptions",
|
| 8 |
"0.5.1": "update the TensorRT part in the README file",
|
| 9 |
"0.5.0": "add the command of executing inference with TensorRT models",
|
|
|
|
| 44 |
"label_classes": "binary labels for each patch",
|
| 45 |
"pred_classes": "scalar probability",
|
| 46 |
"eval_metrics": {
|
| 47 |
+
"accuracy": 0.9,
|
| 48 |
+
"froc": 0.72
|
| 49 |
},
|
| 50 |
"intended_use": "This is an example, not to be used for diagnostic purposes",
|
| 51 |
"references": [
|
configs/multi_gpu_train.json
CHANGED
|
@@ -28,8 +28,7 @@
|
|
| 28 |
"$import torch.distributed as dist",
|
| 29 |
"$dist.is_initialized() or dist.init_process_group(backend='nccl')",
|
| 30 |
"$torch.cuda.set_device(@device)",
|
| 31 |
-
"$monai.utils.set_determinism(seed=123)"
|
| 32 |
-
"$setattr(torch.backends.cudnn, 'benchmark', True)"
|
| 33 |
],
|
| 34 |
"run": [
|
| 35 |
"$@train#trainer.run()"
|
|
|
|
| 28 |
"$import torch.distributed as dist",
|
| 29 |
"$dist.is_initialized() or dist.init_process_group(backend='nccl')",
|
| 30 |
"$torch.cuda.set_device(@device)",
|
| 31 |
+
"$monai.utils.set_determinism(seed=123)"
|
|
|
|
| 32 |
],
|
| 33 |
"run": [
|
| 34 |
"$@train#trainer.run()"
|
configs/train.json
CHANGED
|
@@ -372,8 +372,7 @@
|
|
| 372 |
}
|
| 373 |
},
|
| 374 |
"initialize": [
|
| 375 |
-
"$monai.utils.set_determinism(seed=15)"
|
| 376 |
-
"$setattr(torch.backends.cudnn, 'benchmark', True)"
|
| 377 |
],
|
| 378 |
"run": [
|
| 379 |
"$@train#trainer.run()"
|
|
|
|
| 372 |
}
|
| 373 |
},
|
| 374 |
"initialize": [
|
| 375 |
+
"$monai.utils.set_determinism(seed=15)"
|
|
|
|
| 376 |
],
|
| 377 |
"run": [
|
| 378 |
"$@train#trainer.run()"
|
docs/README.md
CHANGED
|
@@ -39,6 +39,21 @@ The training was performed with the following:
|
|
| 39 |
- Loss: BCEWithLogitsLoss
|
| 40 |
- Whole slide image reader: cuCIM (if running on Windows or Mac, please install `OpenSlide` on your system and change `wsi_reader` to "OpenSlide")
|
| 41 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 42 |
### Input
|
| 43 |
|
| 44 |
The training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
|
|
@@ -51,12 +66,16 @@ A probability number of the input patch being tumor or normal.
|
|
| 51 |
|
| 52 |
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
|
| 53 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 54 |
## Performance
|
| 55 |
|
| 56 |
FROC score is used for evaluating the performance of the model. After inference is done, `evaluate_froc.sh` needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
|
| 57 |
-
|
| 58 |
|
| 59 |
-

|
| 41 |
|
| 42 |
+
### Pretrained Weights
|
| 43 |
+
|
| 44 |
+
By setting the `"pretrained"` parameter of `TorchVisionFCModel` in the config file to `true`, ImageNet pre-trained weights will be used for training. Please note that these weights are for non-commercial use. Each user is responsible for checking the content of the models/datasets and the applicable licenses and determining if suitable for the intended use. In order to use other pretrained weights, you can use `CheckpointLoader` in train handlers section as the first handler:
|
| 45 |
+
|
| 46 |
+
```json
|
| 47 |
+
{
|
| 48 |
+
"_target_": "CheckpointLoader",
|
| 49 |
+
"load_path": "$@bundle_root + '/pretrained_resnet18.pth'",
|
| 50 |
+
"strict": false,
|
| 51 |
+
"load_dict": {
|
| 52 |
+
"model_new": "@network"
|
| 53 |
+
}
|
| 54 |
+
}
|
| 55 |
+
```
|
| 56 |
+
|
| 57 |
### Input
|
| 58 |
|
| 59 |
The training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
|
|
|
|
| 66 |
|
| 67 |
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
|
| 68 |
|
| 69 |
+
### Note on determinism
|
| 70 |
+
|
| 71 |
+
By default this bundle use a deterministic approach to make the results reproducible. However, it comes at a cost of performance loss. Thus if you do not care about reproducibility, you can have a performance gain by replacing `"$monai.utils.set_determinism"` line with `"$setattr(torch.backends.cudnn, 'benchmark', True)"` in initialize section of training configuration (`configs/train.json` and `configs/multi_gpu_train.json` for single GPU and multi-GPU training respectively).
|
| 72 |
+
|
| 73 |
## Performance
|
| 74 |
|
| 75 |
FROC score is used for evaluating the performance of the model. After inference is done, `evaluate_froc.sh` needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
|
| 76 |
+
Using an internal pretrained weights for ResNet18, this model deterministically achieves the 0.90 accuracy on validation patches, and FROC of 0.72 on the 48 Camelyon testing data that have ground truth annotations available.
|
| 77 |
|
| 78 |
+

|
| 79 |
|
| 80 |
The `pathology_tumor_detection` bundle supports acceleration with TensorRT. The table below displays the speedup ratios observed on an A100 80G GPU.
|
| 81 |
|
|
|
|
| 87 |
| end2end |224.97 | 223.50 | 222.65 | 224.03 | 1.01 | 1.01 | 1.00 | 1.00 |
|
| 88 |
|
| 89 |
Where:
|
| 90 |
+
|
| 91 |
- `model computation` means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
|
| 92 |
- `end2end` means run the bundle end-to-end with the TensorRT based model.
|
| 93 |
- `torch_fp32` and `torch_amp` are for the PyTorch models with or without `amp` mode.
|
models/model.pt
CHANGED
|
@@ -1,3 +1,3 @@
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
-
oid sha256:
|
| 3 |
size 44780565
|
|
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:5a0d9b9e714e18a90c1f7f7d9c7e47f807c59f9f8c681b84865fae208fcbb4d6
|
| 3 |
size 44780565
|