Update figure and benchmarking
Browse files- README.md +14 -3
- configs/metadata.json +2 -1
- docs/README.md +14 -3
README.md
CHANGED
|
@@ -10,6 +10,11 @@ A pre-trained model for volumetric (3D) multi-organ segmentation from CT image.
|
|
| 10 |
|
| 11 |
# Model Overview
|
| 12 |
A pre-trained Swin UNETR [1,2] for volumetric (3D) multi-organ segmentation using CT images from Beyond the Cranial Vault (BTCV) Segmentation Challenge dataset [3].
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 13 |
## Data
|
| 14 |
The training data is from the [BTCV dataset](https://www.synapse.org/#!Synapse:syn3193805/wiki/89480/) (Please regist in `Synapse` and download the `Abdomen/RawData.zip`).
|
| 15 |
The dataset format needs to be redefined using the following commands:
|
|
@@ -34,16 +39,22 @@ Actual Model Input: 96 x 96 x 96
|
|
| 34 |
## Input and output formats
|
| 35 |
Input: 1 channel CT image
|
| 36 |
|
| 37 |
-
Output: 14 channels: 0:Background, 1:Spleen, 2:Right Kidney, 3:Left
|
| 38 |
|
| 39 |
## Performance
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 40 |
A graph showing the validation mean Dice for 5000 epochs.
|
| 41 |
|
| 42 |
-
|
| 43 |
|
| 44 |
This model achieves the following Dice score on the validation data (our own split from the training dataset):
|
| 45 |
|
| 46 |
-
Mean Dice = 0.
|
| 47 |
|
| 48 |
Note that mean dice is computed in the original spacing of the input data.
|
| 49 |
## commands example
|
|
|
|
| 10 |
|
| 11 |
# Model Overview
|
| 12 |
A pre-trained Swin UNETR [1,2] for volumetric (3D) multi-organ segmentation using CT images from Beyond the Cranial Vault (BTCV) Segmentation Challenge dataset [3].
|
| 13 |
+
|
| 14 |
+
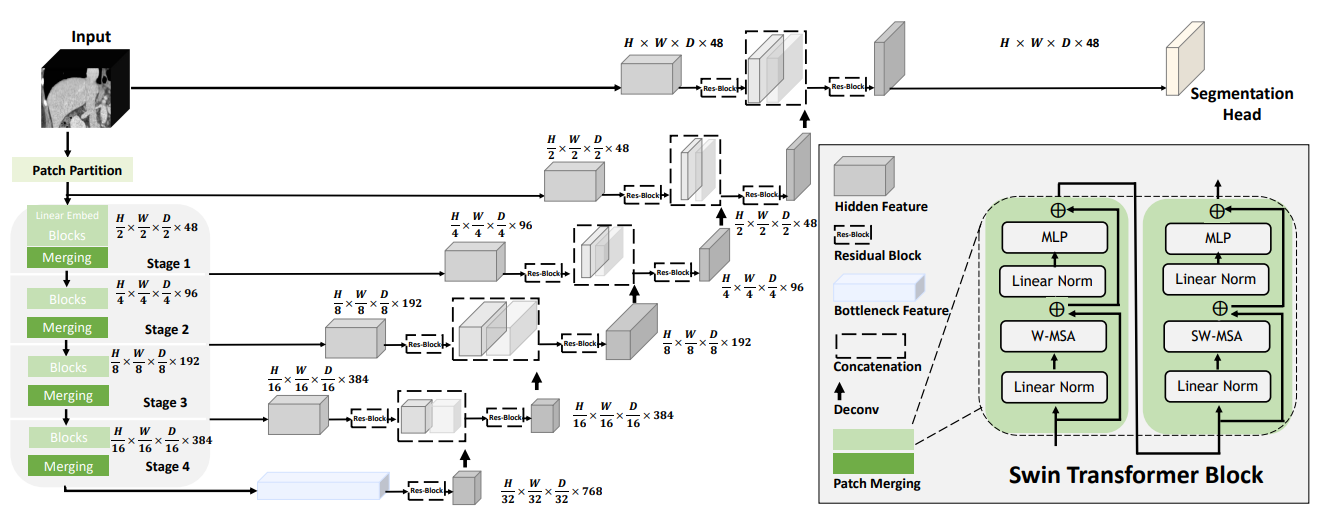
The architecture of Swin UNETR is shown as below:
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
|
| 18 |
## Data
|
| 19 |
The training data is from the [BTCV dataset](https://www.synapse.org/#!Synapse:syn3193805/wiki/89480/) (Please regist in `Synapse` and download the `Abdomen/RawData.zip`).
|
| 20 |
The dataset format needs to be redefined using the following commands:
|
|
|
|
| 39 |
## Input and output formats
|
| 40 |
Input: 1 channel CT image
|
| 41 |
|
| 42 |
+
Output: 14 channels: 0:Background, 1:Spleen, 2:Right Kidney, 3:Left Kidney, 4:Gallbladder, 5:Esophagus, 6:Liver, 7:Stomach, 8:Aorta, 9:IVC, 10:Portal and Splenic Veins, 11:Pancreas, 12:Right adrenal gland, 13:Left adrenal gland
|
| 43 |
|
| 44 |
## Performance
|
| 45 |
+
|
| 46 |
+
The figure shows the training loss curve for 10K iterations.
|
| 47 |
+
|
| 48 |
+
|
| 49 |
+
<p align = "center"><img src="https://developer.download.nvidia.com/assets/Clara/Images/monai_swin_unetr_btcv_segmentation_trainloss_v1.png" alt="drawing" width="700"/></p>
|
| 50 |
+
|
| 51 |
A graph showing the validation mean Dice for 5000 epochs.
|
| 52 |
|
| 53 |
+
<p align = "center"><img src="https://developer.download.nvidia.com/assets/Clara/Images/monai_swin_unetr_btcv_segmentation_validation_meandice_v1.png" alt="drawing" width="700"/></p>
|
| 54 |
|
| 55 |
This model achieves the following Dice score on the validation data (our own split from the training dataset):
|
| 56 |
|
| 57 |
+
Mean Dice = 0.8269
|
| 58 |
|
| 59 |
Note that mean dice is computed in the original spacing of the input data.
|
| 60 |
## commands example
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,8 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.3.
|
| 4 |
"changelog": {
|
|
|
|
| 5 |
"0.3.4": "Update figure link in readme",
|
| 6 |
"0.3.3": "Update, verify MONAI 1.0.1 and Pytorch 1.13.0",
|
| 7 |
"0.3.2": "enhance readme on commands example",
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.3.5",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.3.5": "Update figure and benchmarking",
|
| 6 |
"0.3.4": "Update figure link in readme",
|
| 7 |
"0.3.3": "Update, verify MONAI 1.0.1 and Pytorch 1.13.0",
|
| 8 |
"0.3.2": "enhance readme on commands example",
|
docs/README.md
CHANGED
|
@@ -3,6 +3,11 @@ A pre-trained model for volumetric (3D) multi-organ segmentation from CT image.
|
|
| 3 |
|
| 4 |
# Model Overview
|
| 5 |
A pre-trained Swin UNETR [1,2] for volumetric (3D) multi-organ segmentation using CT images from Beyond the Cranial Vault (BTCV) Segmentation Challenge dataset [3].
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 6 |
## Data
|
| 7 |
The training data is from the [BTCV dataset](https://www.synapse.org/#!Synapse:syn3193805/wiki/89480/) (Please regist in `Synapse` and download the `Abdomen/RawData.zip`).
|
| 8 |
The dataset format needs to be redefined using the following commands:
|
|
@@ -27,16 +32,22 @@ Actual Model Input: 96 x 96 x 96
|
|
| 27 |
## Input and output formats
|
| 28 |
Input: 1 channel CT image
|
| 29 |
|
| 30 |
-
Output: 14 channels: 0:Background, 1:Spleen, 2:Right Kidney, 3:Left
|
| 31 |
|
| 32 |
## Performance
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 33 |
A graph showing the validation mean Dice for 5000 epochs.
|
| 34 |
|
| 35 |
-
|
| 36 |
|
| 37 |
This model achieves the following Dice score on the validation data (our own split from the training dataset):
|
| 38 |
|
| 39 |
-
Mean Dice = 0.
|
| 40 |
|
| 41 |
Note that mean dice is computed in the original spacing of the input data.
|
| 42 |
## commands example
|
|
|
|
| 3 |
|
| 4 |
# Model Overview
|
| 5 |
A pre-trained Swin UNETR [1,2] for volumetric (3D) multi-organ segmentation using CT images from Beyond the Cranial Vault (BTCV) Segmentation Challenge dataset [3].
|
| 6 |
+
|
| 7 |
+
The architecture of Swin UNETR is shown as below:
|
| 8 |
+
|
| 9 |
+

|
| 10 |
+
|
| 11 |
## Data
|
| 12 |
The training data is from the [BTCV dataset](https://www.synapse.org/#!Synapse:syn3193805/wiki/89480/) (Please regist in `Synapse` and download the `Abdomen/RawData.zip`).
|
| 13 |
The dataset format needs to be redefined using the following commands:
|
|
|
|
| 32 |
## Input and output formats
|
| 33 |
Input: 1 channel CT image
|
| 34 |
|
| 35 |
+
Output: 14 channels: 0:Background, 1:Spleen, 2:Right Kidney, 3:Left Kidney, 4:Gallbladder, 5:Esophagus, 6:Liver, 7:Stomach, 8:Aorta, 9:IVC, 10:Portal and Splenic Veins, 11:Pancreas, 12:Right adrenal gland, 13:Left adrenal gland
|
| 36 |
|
| 37 |
## Performance
|
| 38 |
+
|
| 39 |
+
The figure shows the training loss curve for 10K iterations.
|
| 40 |
+
|
| 41 |
+
|
| 42 |
+
<p align = "center"><img src="https://developer.download.nvidia.com/assets/Clara/Images/monai_swin_unetr_btcv_segmentation_trainloss_v1.png" alt="drawing" width="700"/></p>
|
| 43 |
+
|
| 44 |
A graph showing the validation mean Dice for 5000 epochs.
|
| 45 |
|
| 46 |
+
<p align = "center"><img src="https://developer.download.nvidia.com/assets/Clara/Images/monai_swin_unetr_btcv_segmentation_validation_meandice_v1.png" alt="drawing" width="700"/></p>
|
| 47 |
|
| 48 |
This model achieves the following Dice score on the validation data (our own split from the training dataset):
|
| 49 |
|
| 50 |
+
Mean Dice = 0.8269
|
| 51 |
|
| 52 |
Note that mean dice is computed in the original spacing of the input data.
|
| 53 |
## commands example
|