Spaces:
Running
Running
A newer version of the Streamlit SDK is available:
1.42.0
metadata
title: Ontograph
emoji: 💻
colorFrom: red
colorTo: green
sdk: streamlit
sdk_version: 1.41.1
app_file: app.py
pinned: false
🌟 OntoGraph - Drug Interaction Analysis System
A sophisticated Python-based application that combines ontology reasoning with LLM capabilities to analyze drug interactions, conflicts, and alternatives. The system provides comprehensive analysis through both CLI and web interfaces.
🚀 Key Features
- Dual Interface Options:
- Web-based interface using Streamlit
- Command-line interface for direct interaction
- Comprehensive Drug Analysis:
- Drug interaction detection
- Conflict identification
- Similar drug suggestions
- Alternative medication recommendations
- Advanced Technology Stack:
- RDF/OWL ontology for knowledge representation
- Groq LLM integration for natural language processing
- Agent-based architecture using Langgraph for modular analysis
- Structured logging system
🛠️ Technical Requirements
- Python 3.x
- Groq API key
- Required Python packages:
- rdflib
- langchain
- python-dotenv
- langchain-community
- langchain-groq
- langgraph
- streamlit
📥 Installation
- Clone the repository:
git clone https://github.com/Manithj/onto_graph.git
cd onto_graph
- Install dependencies:
pip install -r requirements.txt
- Configure environment:
- Create a
.envfile in the project root - Add your Groq API key:
- Create a
GROQ_API_KEY=your_api_key_here
💻 Usage
Web Interface
streamlit run app.py
Navigate to the displayed local URL to access the web interface.
Command Line Interface
python appcli.py
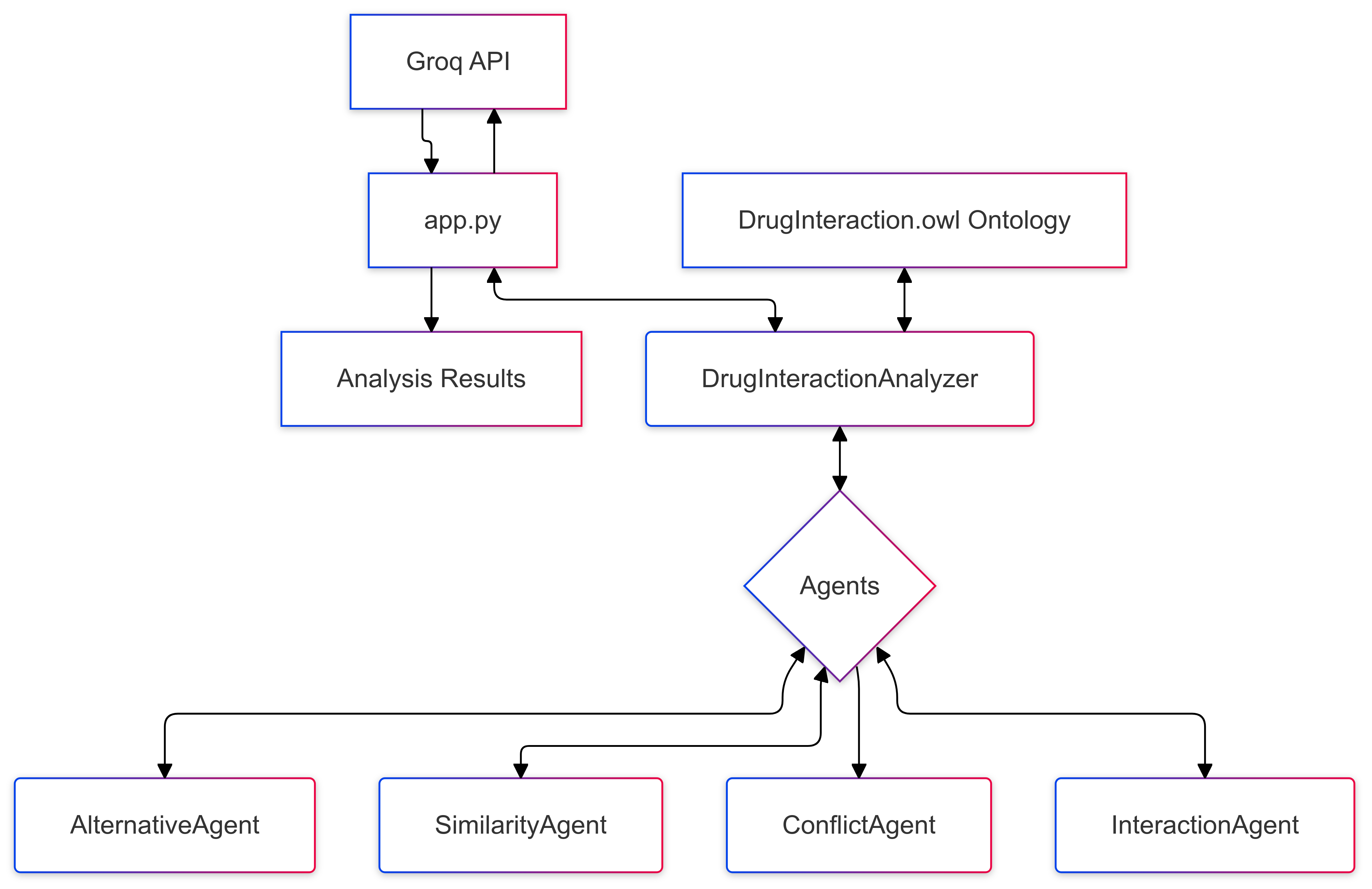
🏗️ Project Structure
onto_graph/
├── app.py # Streamlit web interface
├── appcli.py # Command line interface
├── agents/ # Agent implementations
│ ├── __init__.py
│ ├── alternative_agent.py
│ ├── base_agent.py
│ ├── conflict_agent.py
│ ├── interaction_agent.py
│ └── similarity_agent.py
├── analyzers/ # Analysis coordination
│ ├── __init__.py
│ └── drug_interaction_analyzer.py
├── ontology/ # Knowledge base
│ └── DrugInteraction.owl
└── requirements.txt # Project dependencies
🔍 Features in Detail
Agent System
- Base Agent: Common functionality for ontology queries
- Alternative Agent: Identifies alternative medications
- Similarity Agent: Finds similar drugs
- Conflict Agent: Detects drug conflicts
- Interaction Agent: Analyzes drug interactions
Analysis Pipeline
- User input processing
- Ontology querying
- LLM-powered result synthesis
- Structured response generation
🤝 Contributing
We welcome contributions! Please follow these steps:
- Fork the repository
- Create a feature branch
- Commit your changes
- Push to your branch
- Create a Pull Request
📝 Logging
The system maintains detailed logs in app.log, capturing:
- Information level messages
- Warning and error states
- Query execution details
- System state changes
📜 License
This project is licensed under the Apache 2.0.