Spaces:
Running
title: DocumentIQA
emoji: 🚀
colorFrom: yellow
colorTo: pink
sdk: streamlit
sdk_version: 1.27.2
app_file: streamlit_app.py
pinned: false
license: apache-2.0
DocumentIQA: Scientific Document Insight QA
Introduction
Question/Answering on scientific documents using LLMs (OpenAI, Mistral, LLama2, etc..).
This application is the frontend for testing the RAG (Retrieval Augmented Generation) on scientific documents, that we are developing at NIMS.
Differently to most of the project, we focus on scientific articles and we are using Grobid for text extraction instead of the raw PDF2Text converter (which is comparable with most of other solutions) allow to extract only full-text.
Work in progress
Demos:
- (on HuggingFace spaces): https://lfoppiano-document-qa.hf.space/
- (on the Streamlit cloud): https://document-insights.streamlit.app/
Getting started
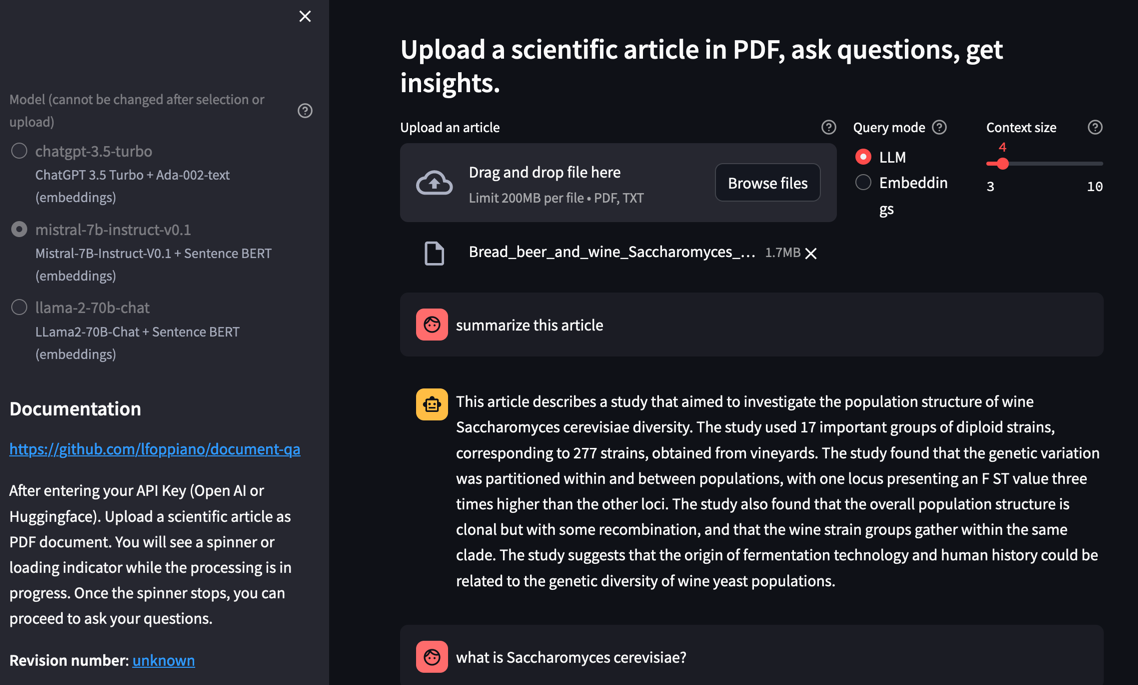
- Select the model+embedding combination you want ot use
(for LLama2 you must acknowledge their licence both on meta.com and on huggingface. See here)(Llama2 was removed due to API limitations). - Enter your API Key (Open AI or Huggingface).
- Upload a scientific article as PDF document. You will see a spinner or loading indicator while the processing is in progress.
- Once the spinner stops, you can proceed to ask your questions
Options
Context size
Allow to change the number of embedding chunks that are considered for responding. The text chunk are around 250 tokens, which uses around 1000 tokens for each question.
Query mode
By default, the mode is set to LLM (Language Model) which enables question/answering. You can directly ask questions related to the document content, and the system will answer the question using content from the document. If you switch the mode to "Embedding," the system will return specific chunks from the document that are semantically related to your query. This mode helps to test why sometimes the answers are not satisfying or incomplete.
Acknolwedgement
This project is developed at the National Institute for Materials Science (NIMS) in Japan in collaboration with the Lambard-ML-Team.