license: cc-by-nc-sa-4.0
tags:
- Helical
- RNA
- Transformers
- Sequence
- biology
- mrna
- rna
- genomics
library_name: transformers
Helix-mRNA-v0
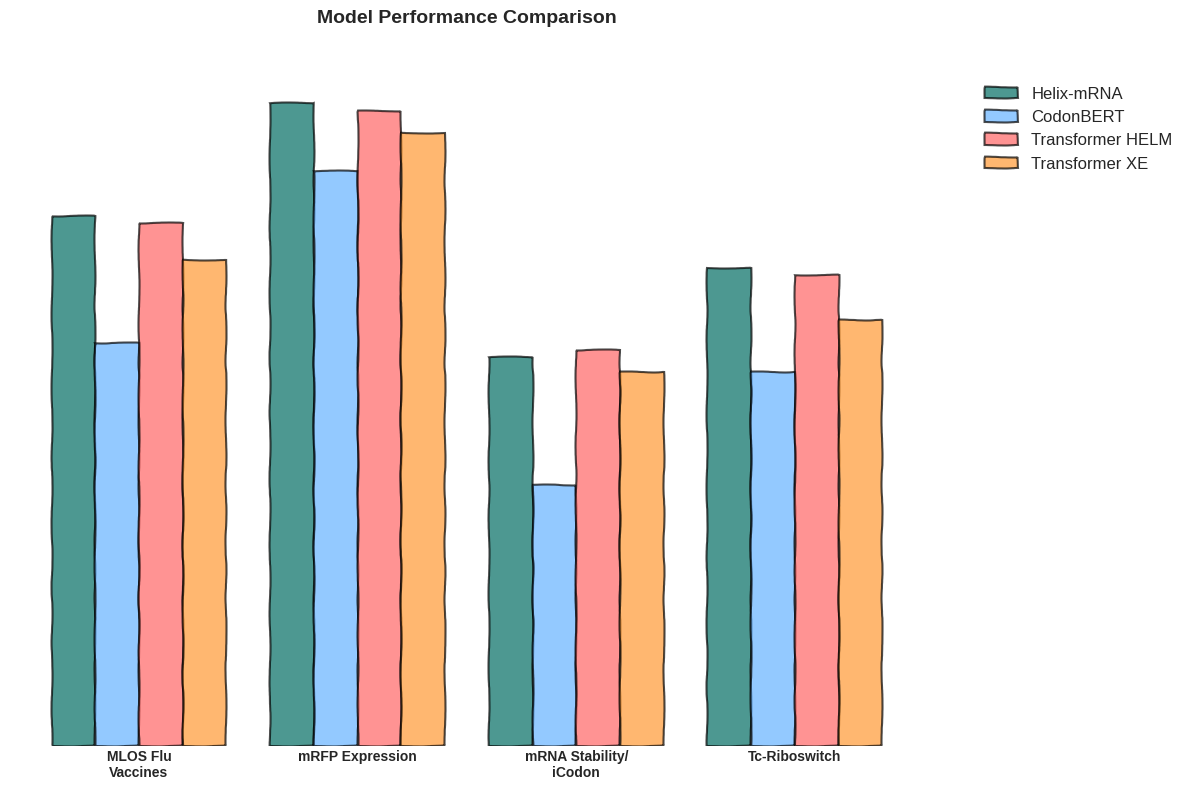
Helix-mRNA emerges as a hybrid state-space and transformer based model, leveraging both the efficient sequence processing capabilities of Mamba2's state-space architecture and the contextual understanding of transformer attention mechanisms, allowing for the best of both worlds between these two approaches. These traits make it particularly suitable for studying full-length transcripts, splice variants, and complex mRNA structural elements.
We tokenize mRNA sequences at single-nucleotide resolution by mapping each nucleotide (A, C, U, G) and ambiguous base (N) to a unique integer. A further special character E is incorporated into the sequence, denoting the start of each codon. This fine-grained approach maximizes the model's ability to extract patterns from the sequences. Unlike coarser tokenization methods that might group nucleotides together or use k-mer based approaches, our single-nucleotide resolution preserves the full sequential information of the mRNA molecule. This simple yet effective encoding scheme ensures that no information is lost during the preprocessing stage, allowing the downstream model to learn directly from the raw sequence composition.
Read more about it in our blog post!
Helical
Install the package
Run the following to install the Helical package via pip:
pip install --upgrade helical
Generate Embeddings
from helical import HelixmRNA, HelixmRNAConfig
import torch
device = "cuda" if torch.cuda.is_available() else "cpu"
input_sequences = ["EACU"*20, "EAUG"*20, "EAUG"*20, "EACU"*20, "EAUU"*20]
helix_mrna_config = HelixmRNAConfig(batch_size=5, device=device, max_length=100)
helix_mrna = HelixmRNA(configurer=helix_mrna_config)
# prepare data for input to the model
processed_input_data = helix_mrna.process_data(input_sequences)
# generate the embeddings for the processed data
embeddings = helix_mrna.get_embeddings(processed_input_data)
Fine-Tuning
Classification fine-tuning example:
from helical import HelixmRNAFineTuningModel, HelixmRNAConfig
import torch
device = "cuda" if torch.cuda.is_available() else "cpu"
input_sequences = ["EACU"*20, "EAUG"*20, "EAUG"*20, "EACU"*20, "EAUU"*20]
labels = [0, 2, 2, 0, 1]
helixr_config = HelixmRNAConfig(batch_size=5, device=device, max_length=100)
helixr_fine_tune = HelixmRNAFineTuningModel(helix_mrna_config=helixr_config, fine_tuning_head="classification", output_size=3)
# prepare data for input to the model
train_dataset = helixr_fine_tune.process_data(input_sequences)
# fine-tune the model with the relevant training labels
helixr_fine_tune.train(train_dataset=train_dataset, train_labels=labels)
# get outputs from the fine-tuned model on a processed dataset
outputs = helixr_fine_tune.get_outputs(train_dataset)
@software{allard_2024_13135902,
author = {Helical Team},
title = {helicalAI/helical: v0.0.1-alpha10},
month = nov,
year = 2024,
publisher = {Zenodo},
version = {0.0.1a10},
doi = {10.5281/zenodo.13135902},
url = {https://doi.org/10.5281/zenodo.13135902}
}