tags:
- monai
- medical
library_name: monai
license: unknown
Model Overview
A pre-trained model for automated detection of metastases in whole-slide histopathology images.
Workflow
The model is trained based on ResNet18 [1] with the last fully connected layer replaced by a 1x1 convolution layer.
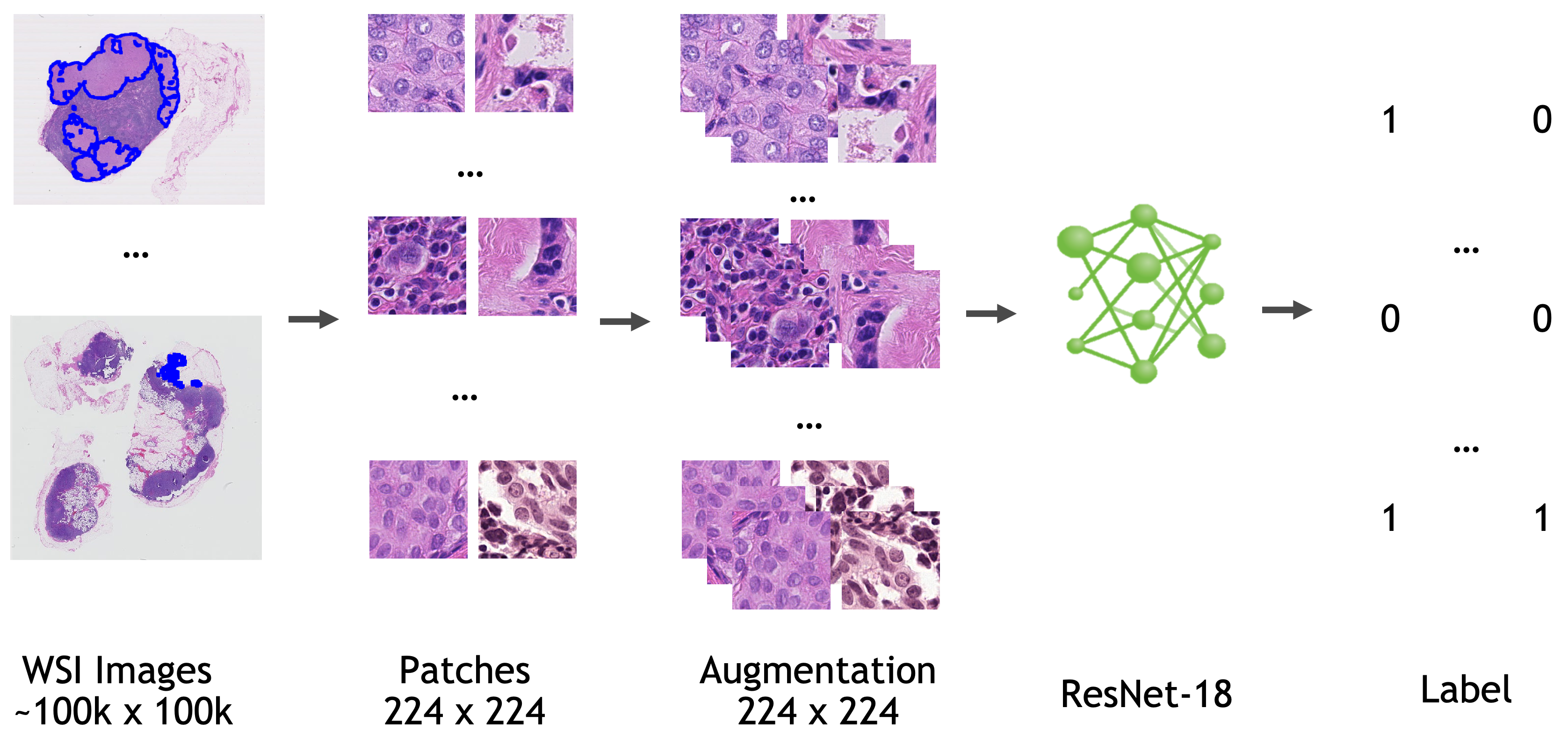
Data
All the data used to train, validate, and test this model is from Camelyon-16 Challenge. You can download all the images for "CAMELYON16" data set from various sources listed here.
Location information for training/validation patches (the location on the whole slide image where patches are extracted) are adopted from NCRF/coords.
Annotation information are adopted from NCRF/jsons.
- Target: Tumor
- Task: Detection
- Modality: Histopathology
- Size: 270 WSIs for training/validation, 48 WSIs for testing
Data Preparation
This MMAR expects the training/validation data (whole slide images) reside in $DATA_ROOT/training/images. By default $DATA_ROOT is pointing to /workspace/data/medical/pathology/ You can easily modify $DATA_ROOT to point to a different directory in config/environment.json.
To reduce the computation burden during the inference, patches are extracted only where there is tissue and ignoring the background according to a tissue mask. You should run prepare_inference_data.sh prior to the inference to generate foreground masks, where the input is the whole slide test images and the output is the foreground masks. Please also create a directory for prediction output, aligning with the one specified with $MMAR_EVAL_OUTPUT_PATH in config/environment.json (e.g. /eval)
Please refer to "Annotation" section of Camelyon challenge to prepare ground truth images, which are needed for FROC computation. By default, this data set is expected to be at /workspace/data/medical/pathology/ground_truths. But it can be modified in evaluate_froc.sh.
Training configuration
The training was performed with the following:
- Script: train.sh
- GPU: at least 16 GB of GPU memory.
- Actual Model Input: 224 x 224 x 3
- AMP: True
- Optimizer: Novograd
- Learning Rate: 1e-3
- Loss: BCEWithLogitsLoss
Input
Input: Input for the training pipeline is a json file (dataset.json) which includes path to each WSI, the location and the label information for each training patch.
- Extract 224 x 224 x 3 patch from WSI according to the location information from json
- Randomly applying color jittering
- Randomly applying spatial flipping
- Randomly applying spatial rotation
- Randomly applying spatial zooming
- Randomly applying intensity scaling
Output
Output of the network is a probability number of the input patch being tumor or normal.
Inference on a WSI
Inference is performed on WSI in a sliding window manner with specified stride. A foreground mask is needed to specify the region where the inference will be performed on, given that background region which contains no tissue at all can occupy a significant portion of a WSI. Output of the inference pipeline is a probability map of size 1/stride of original WSI size.
Model Performance
FROC score is used for evaluating the performance of the model. After inference is done, evaluate_froc.sh needs to be run to evaluate FROC score based on predicted probability map (output of inference) and the ground truth tumor masks.
This model achieve the ~0.92 accuracy on validation patches, and FROC of ~0.72 on the 48 Camelyon testing data that have ground truth annotations available.
Commands example
Execute training:
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.json --logging_file configs/logging.conf
Override the train config to execute multi-GPU training:
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.json','configs/multi_gpu_train.json']" --logging_file configs/logging.conf
Override the train config to execute evaluation with the trained model:
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.json','configs/evaluate.json']" --logging_file configs/logging.conf
Execute inference:
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.json --logging_file configs/logging.conf
Intended Use
The model needs to be used with NVIDIA hardware and software. For hardware, the model can run on any NVIDIA GPU with memory greater than 16 GB. For software, this model is usable only as part of Transfer Learning & Annotation Tools in Clara Train SDK container. Find out more about Clara Train at the Clara Train Collections on NGC.
The pre-trained models are for developmental purposes only and cannot be used directly for clinical procedures.
License
End User License Agreement is included with the product. Licenses are also available along with the model application zip file. By pulling and using the Clara Train SDK container and downloading models, you accept the terms and conditions of these licenses.
References
[1] He, Kaiming, et al, "Deep Residual Learning for Image Recognition." In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770-778. 2016. https://arxiv.org/pdf/1512.03385.pdf