repo_name
stringlengths 7
92
| path
stringlengths 5
149
| copies
stringlengths 1
3
| size
stringlengths 4
6
| content
stringlengths 911
693k
| license
stringclasses 15
values |
|---|---|---|---|---|---|
nomadcube/scikit-learn | examples/mixture/plot_gmm_pdf.py | 284 | 1528 | """
=============================================
Density Estimation for a mixture of Gaussians
=============================================
Plot the density estimation of a mixture of two Gaussians. Data is
generated from two Gaussians with different centers and covariance
matrices.
"""
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
from sklearn import mixture
n_samples = 300
# generate random sample, two components
np.random.seed(0)
# generate spherical data centered on (20, 20)
shifted_gaussian = np.random.randn(n_samples, 2) + np.array([20, 20])
# generate zero centered stretched Gaussian data
C = np.array([[0., -0.7], [3.5, .7]])
stretched_gaussian = np.dot(np.random.randn(n_samples, 2), C)
# concatenate the two datasets into the final training set
X_train = np.vstack([shifted_gaussian, stretched_gaussian])
# fit a Gaussian Mixture Model with two components
clf = mixture.GMM(n_components=2, covariance_type='full')
clf.fit(X_train)
# display predicted scores by the model as a contour plot
x = np.linspace(-20.0, 30.0)
y = np.linspace(-20.0, 40.0)
X, Y = np.meshgrid(x, y)
XX = np.array([X.ravel(), Y.ravel()]).T
Z = -clf.score_samples(XX)[0]
Z = Z.reshape(X.shape)
CS = plt.contour(X, Y, Z, norm=LogNorm(vmin=1.0, vmax=1000.0),
levels=np.logspace(0, 3, 10))
CB = plt.colorbar(CS, shrink=0.8, extend='both')
plt.scatter(X_train[:, 0], X_train[:, 1], .8)
plt.title('Negative log-likelihood predicted by a GMM')
plt.axis('tight')
plt.show()
| bsd-3-clause |
mikelseverson/Udacity-Deep_Learning-Nanodegree | weight-initialization/helper.py | 153 | 3649 | import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
def hist_dist(title, distribution_tensor, hist_range=(-4, 4)):
"""
Display histogram of a TF distribution
"""
with tf.Session() as sess:
values = sess.run(distribution_tensor)
plt.title(title)
plt.hist(values, np.linspace(*hist_range, num=len(values)/2))
plt.show()
def _get_loss_acc(dataset, weights):
"""
Get losses and validation accuracy of example neural network
"""
batch_size = 128
epochs = 2
learning_rate = 0.001
features = tf.placeholder(tf.float32)
labels = tf.placeholder(tf.float32)
learn_rate = tf.placeholder(tf.float32)
biases = [
tf.Variable(tf.zeros([256])),
tf.Variable(tf.zeros([128])),
tf.Variable(tf.zeros([dataset.train.labels.shape[1]]))
]
# Layers
layer_1 = tf.nn.relu(tf.matmul(features, weights[0]) + biases[0])
layer_2 = tf.nn.relu(tf.matmul(layer_1, weights[1]) + biases[1])
logits = tf.matmul(layer_2, weights[2]) + biases[2]
# Training loss
loss = tf.reduce_mean(tf.nn.softmax_cross_entropy_with_logits(logits=logits, labels=labels))
# Optimizer
optimizer = tf.train.AdamOptimizer(learn_rate).minimize(loss)
# Accuracy
correct_prediction = tf.equal(tf.argmax(logits, 1), tf.argmax(labels, 1))
accuracy = tf.reduce_mean(tf.cast(correct_prediction, tf.float32))
# Measurements use for graphing loss
loss_batch = []
with tf.Session() as session:
session.run(tf.global_variables_initializer())
batch_count = int((dataset.train.num_examples / batch_size))
# The training cycle
for epoch_i in range(epochs):
for batch_i in range(batch_count):
batch_features, batch_labels = dataset.train.next_batch(batch_size)
# Run optimizer and get loss
session.run(
optimizer,
feed_dict={features: batch_features, labels: batch_labels, learn_rate: learning_rate})
l = session.run(
loss,
feed_dict={features: batch_features, labels: batch_labels, learn_rate: learning_rate})
loss_batch.append(l)
valid_acc = session.run(
accuracy,
feed_dict={features: dataset.validation.images, labels: dataset.validation.labels, learn_rate: 1.0})
# Hack to Reset batches
dataset.train._index_in_epoch = 0
dataset.train._epochs_completed = 0
return loss_batch, valid_acc
def compare_init_weights(
dataset,
title,
weight_init_list,
plot_n_batches=100):
"""
Plot loss and print stats of weights using an example neural network
"""
colors = ['r', 'b', 'g', 'c', 'y', 'k']
label_accs = []
label_loss = []
assert len(weight_init_list) <= len(colors), 'Too many inital weights to plot'
for i, (weights, label) in enumerate(weight_init_list):
loss, val_acc = _get_loss_acc(dataset, weights)
plt.plot(loss[:plot_n_batches], colors[i], label=label)
label_accs.append((label, val_acc))
label_loss.append((label, loss[-1]))
plt.title(title)
plt.xlabel('Batches')
plt.ylabel('Loss')
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
plt.show()
print('After 858 Batches (2 Epochs):')
print('Validation Accuracy')
for label, val_acc in label_accs:
print(' {:7.3f}% -- {}'.format(val_acc*100, label))
print('Loss')
for label, loss in label_loss:
print(' {:7.3f} -- {}'.format(loss, label))
| mit |
mhdella/scikit-learn | sklearn/neighbors/tests/test_kd_tree.py | 129 | 7848 | import numpy as np
from numpy.testing import assert_array_almost_equal
from sklearn.neighbors.kd_tree import (KDTree, NeighborsHeap,
simultaneous_sort, kernel_norm,
nodeheap_sort, DTYPE, ITYPE)
from sklearn.neighbors.dist_metrics import DistanceMetric
from sklearn.utils.testing import SkipTest, assert_allclose
V = np.random.random((3, 3))
V = np.dot(V, V.T)
DIMENSION = 3
METRICS = {'euclidean': {},
'manhattan': {},
'chebyshev': {},
'minkowski': dict(p=3)}
def brute_force_neighbors(X, Y, k, metric, **kwargs):
D = DistanceMetric.get_metric(metric, **kwargs).pairwise(Y, X)
ind = np.argsort(D, axis=1)[:, :k]
dist = D[np.arange(Y.shape[0])[:, None], ind]
return dist, ind
def test_kd_tree_query():
np.random.seed(0)
X = np.random.random((40, DIMENSION))
Y = np.random.random((10, DIMENSION))
def check_neighbors(dualtree, breadth_first, k, metric, kwargs):
kdt = KDTree(X, leaf_size=1, metric=metric, **kwargs)
dist1, ind1 = kdt.query(Y, k, dualtree=dualtree,
breadth_first=breadth_first)
dist2, ind2 = brute_force_neighbors(X, Y, k, metric, **kwargs)
# don't check indices here: if there are any duplicate distances,
# the indices may not match. Distances should not have this problem.
assert_array_almost_equal(dist1, dist2)
for (metric, kwargs) in METRICS.items():
for k in (1, 3, 5):
for dualtree in (True, False):
for breadth_first in (True, False):
yield (check_neighbors,
dualtree, breadth_first,
k, metric, kwargs)
def test_kd_tree_query_radius(n_samples=100, n_features=10):
np.random.seed(0)
X = 2 * np.random.random(size=(n_samples, n_features)) - 1
query_pt = np.zeros(n_features, dtype=float)
eps = 1E-15 # roundoff error can cause test to fail
kdt = KDTree(X, leaf_size=5)
rad = np.sqrt(((X - query_pt) ** 2).sum(1))
for r in np.linspace(rad[0], rad[-1], 100):
ind = kdt.query_radius(query_pt, r + eps)[0]
i = np.where(rad <= r + eps)[0]
ind.sort()
i.sort()
assert_array_almost_equal(i, ind)
def test_kd_tree_query_radius_distance(n_samples=100, n_features=10):
np.random.seed(0)
X = 2 * np.random.random(size=(n_samples, n_features)) - 1
query_pt = np.zeros(n_features, dtype=float)
eps = 1E-15 # roundoff error can cause test to fail
kdt = KDTree(X, leaf_size=5)
rad = np.sqrt(((X - query_pt) ** 2).sum(1))
for r in np.linspace(rad[0], rad[-1], 100):
ind, dist = kdt.query_radius(query_pt, r + eps, return_distance=True)
ind = ind[0]
dist = dist[0]
d = np.sqrt(((query_pt - X[ind]) ** 2).sum(1))
assert_array_almost_equal(d, dist)
def compute_kernel_slow(Y, X, kernel, h):
d = np.sqrt(((Y[:, None, :] - X) ** 2).sum(-1))
norm = kernel_norm(h, X.shape[1], kernel)
if kernel == 'gaussian':
return norm * np.exp(-0.5 * (d * d) / (h * h)).sum(-1)
elif kernel == 'tophat':
return norm * (d < h).sum(-1)
elif kernel == 'epanechnikov':
return norm * ((1.0 - (d * d) / (h * h)) * (d < h)).sum(-1)
elif kernel == 'exponential':
return norm * (np.exp(-d / h)).sum(-1)
elif kernel == 'linear':
return norm * ((1 - d / h) * (d < h)).sum(-1)
elif kernel == 'cosine':
return norm * (np.cos(0.5 * np.pi * d / h) * (d < h)).sum(-1)
else:
raise ValueError('kernel not recognized')
def test_kd_tree_kde(n_samples=100, n_features=3):
np.random.seed(0)
X = np.random.random((n_samples, n_features))
Y = np.random.random((n_samples, n_features))
kdt = KDTree(X, leaf_size=10)
for kernel in ['gaussian', 'tophat', 'epanechnikov',
'exponential', 'linear', 'cosine']:
for h in [0.01, 0.1, 1]:
dens_true = compute_kernel_slow(Y, X, kernel, h)
def check_results(kernel, h, atol, rtol, breadth_first):
dens = kdt.kernel_density(Y, h, atol=atol, rtol=rtol,
kernel=kernel,
breadth_first=breadth_first)
assert_allclose(dens, dens_true, atol=atol,
rtol=max(rtol, 1e-7))
for rtol in [0, 1E-5]:
for atol in [1E-6, 1E-2]:
for breadth_first in (True, False):
yield (check_results, kernel, h, atol, rtol,
breadth_first)
def test_gaussian_kde(n_samples=1000):
# Compare gaussian KDE results to scipy.stats.gaussian_kde
from scipy.stats import gaussian_kde
np.random.seed(0)
x_in = np.random.normal(0, 1, n_samples)
x_out = np.linspace(-5, 5, 30)
for h in [0.01, 0.1, 1]:
kdt = KDTree(x_in[:, None])
try:
gkde = gaussian_kde(x_in, bw_method=h / np.std(x_in))
except TypeError:
raise SkipTest("Old scipy, does not accept explicit bandwidth.")
dens_kdt = kdt.kernel_density(x_out[:, None], h) / n_samples
dens_gkde = gkde.evaluate(x_out)
assert_array_almost_equal(dens_kdt, dens_gkde, decimal=3)
def test_kd_tree_two_point(n_samples=100, n_features=3):
np.random.seed(0)
X = np.random.random((n_samples, n_features))
Y = np.random.random((n_samples, n_features))
r = np.linspace(0, 1, 10)
kdt = KDTree(X, leaf_size=10)
D = DistanceMetric.get_metric("euclidean").pairwise(Y, X)
counts_true = [(D <= ri).sum() for ri in r]
def check_two_point(r, dualtree):
counts = kdt.two_point_correlation(Y, r=r, dualtree=dualtree)
assert_array_almost_equal(counts, counts_true)
for dualtree in (True, False):
yield check_two_point, r, dualtree
def test_kd_tree_pickle():
import pickle
np.random.seed(0)
X = np.random.random((10, 3))
kdt1 = KDTree(X, leaf_size=1)
ind1, dist1 = kdt1.query(X)
def check_pickle_protocol(protocol):
s = pickle.dumps(kdt1, protocol=protocol)
kdt2 = pickle.loads(s)
ind2, dist2 = kdt2.query(X)
assert_array_almost_equal(ind1, ind2)
assert_array_almost_equal(dist1, dist2)
for protocol in (0, 1, 2):
yield check_pickle_protocol, protocol
def test_neighbors_heap(n_pts=5, n_nbrs=10):
heap = NeighborsHeap(n_pts, n_nbrs)
for row in range(n_pts):
d_in = np.random.random(2 * n_nbrs).astype(DTYPE)
i_in = np.arange(2 * n_nbrs, dtype=ITYPE)
for d, i in zip(d_in, i_in):
heap.push(row, d, i)
ind = np.argsort(d_in)
d_in = d_in[ind]
i_in = i_in[ind]
d_heap, i_heap = heap.get_arrays(sort=True)
assert_array_almost_equal(d_in[:n_nbrs], d_heap[row])
assert_array_almost_equal(i_in[:n_nbrs], i_heap[row])
def test_node_heap(n_nodes=50):
vals = np.random.random(n_nodes).astype(DTYPE)
i1 = np.argsort(vals)
vals2, i2 = nodeheap_sort(vals)
assert_array_almost_equal(i1, i2)
assert_array_almost_equal(vals[i1], vals2)
def test_simultaneous_sort(n_rows=10, n_pts=201):
dist = np.random.random((n_rows, n_pts)).astype(DTYPE)
ind = (np.arange(n_pts) + np.zeros((n_rows, 1))).astype(ITYPE)
dist2 = dist.copy()
ind2 = ind.copy()
# simultaneous sort rows using function
simultaneous_sort(dist, ind)
# simultaneous sort rows using numpy
i = np.argsort(dist2, axis=1)
row_ind = np.arange(n_rows)[:, None]
dist2 = dist2[row_ind, i]
ind2 = ind2[row_ind, i]
assert_array_almost_equal(dist, dist2)
assert_array_almost_equal(ind, ind2)
| bsd-3-clause |
JPalmerio/GRB_population_code | catalogs/GBM_cat/GBM_Ep_constraint_testing.py | 1 | 1178 | import sys
import platform
if platform.system() == 'Linux':
sys.path.insert(0,'/nethome/palmerio/Dropbox/Plotting_GUI/Src')
elif platform.system() == 'Darwin':
sys.path.insert(0,'/Users/palmerio/Dropbox/Plotting_GUI/Src')
import numpy as np
import matplotlib
import matplotlib.pyplot as plt
import plotting_functions as pf
from matplotlib.transforms import blended_transform_factory
plt.style.use('ggplot')
fig = plt.figure()
ax = fig.add_subplot(111)
root_dir = '/nethome/palmerio/1ere_annee/Frederic/GRB_population_code/Model_outputs/'
filename = root_dir +'run_LIA/EpGBM_constraint.dat'
Ep_bins = pf.read_data(filename, 0)
Ep_hist_mod = pf.read_data(filename, 1)
Ep_hist_obs = pf.read_data(filename, 2)
x=np.linspace(1.,4., 500)
y = max(Ep_hist_obs) * pf.gaussian(x, 2.25, 0.35)
y2 = max(Ep_hist_obs) * pf.gaussian(x, 2.25, 0.375)
ep = np.linspace(1,4, 100)
ep_gauss = pf.gaussian(ep, 2.2, 0.4)*max(Ep_hist_obs)
ax.plot(Ep_bins, Ep_hist_obs, label = 'Observations')
#ax.plot(Ep_bins, Ep_hist_mod, label = 'MC simulation')
#ax.plot(ep, ep_gauss, ls=':', lw=2)
ax.plot(x,y, label='gaussian')
ax.plot(x,y2, label='gaussian2')
ax.legend(loc='best')
plt.show()
| gpl-3.0 |
sjperkins/tensorflow | tensorflow/contrib/learn/python/learn/learn_io/data_feeder.py | 88 | 31139 | # Copyright 2016 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Implementations of different data feeders to provide data for TF trainer."""
# TODO(ipolosukhin): Replace this module with feed-dict queue runners & queues.
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import itertools
import math
import numpy as np
import six
from six.moves import xrange # pylint: disable=redefined-builtin
from tensorflow.python.framework import dtypes
from tensorflow.python.framework import ops
from tensorflow.python.ops import array_ops
from tensorflow.python.platform import tf_logging as logging
# pylint: disable=g-multiple-import,g-bad-import-order
from .pandas_io import HAS_PANDAS, extract_pandas_data, extract_pandas_matrix, extract_pandas_labels
from .dask_io import HAS_DASK, extract_dask_data, extract_dask_labels
# pylint: enable=g-multiple-import,g-bad-import-order
def _get_in_out_shape(x_shape, y_shape, n_classes, batch_size=None):
"""Returns shape for input and output of the data feeder."""
x_is_dict, y_is_dict = isinstance(
x_shape, dict), y_shape is not None and isinstance(y_shape, dict)
if y_is_dict and n_classes is not None:
assert (isinstance(n_classes, dict))
if batch_size is None:
batch_size = list(x_shape.values())[0][0] if x_is_dict else x_shape[0]
elif batch_size <= 0:
raise ValueError('Invalid batch_size %d.' % batch_size)
if x_is_dict:
input_shape = {}
for k, v in list(x_shape.items()):
input_shape[k] = [batch_size] + (list(v[1:]) if len(v) > 1 else [1])
else:
x_shape = list(x_shape[1:]) if len(x_shape) > 1 else [1]
input_shape = [batch_size] + x_shape
if y_shape is None:
return input_shape, None, batch_size
def out_el_shape(out_shape, num_classes):
out_shape = list(out_shape[1:]) if len(out_shape) > 1 else []
# Skip first dimension if it is 1.
if out_shape and out_shape[0] == 1:
out_shape = out_shape[1:]
if num_classes is not None and num_classes > 1:
return [batch_size] + out_shape + [num_classes]
else:
return [batch_size] + out_shape
if not y_is_dict:
output_shape = out_el_shape(y_shape, n_classes)
else:
output_shape = dict([
(k, out_el_shape(v, n_classes[k]
if n_classes is not None and k in n_classes else None))
for k, v in list(y_shape.items())
])
return input_shape, output_shape, batch_size
def _data_type_filter(x, y):
"""Filter data types into acceptable format."""
if HAS_DASK:
x = extract_dask_data(x)
if y is not None:
y = extract_dask_labels(y)
if HAS_PANDAS:
x = extract_pandas_data(x)
if y is not None:
y = extract_pandas_labels(y)
return x, y
def _is_iterable(x):
return hasattr(x, 'next') or hasattr(x, '__next__')
def setup_train_data_feeder(x,
y,
n_classes,
batch_size=None,
shuffle=True,
epochs=None):
"""Create data feeder, to sample inputs from dataset.
If `x` and `y` are iterators, use `StreamingDataFeeder`.
Args:
x: numpy, pandas or Dask matrix or dictionary of aforementioned. Also
supports iterables.
y: numpy, pandas or Dask array or dictionary of aforementioned. Also
supports
iterables.
n_classes: number of classes. Must be None or same type as y. In case, `y`
is `dict`
(or iterable which returns dict) such that `n_classes[key] = n_classes for
y[key]`
batch_size: size to split data into parts. Must be >= 1.
shuffle: Whether to shuffle the inputs.
epochs: Number of epochs to run.
Returns:
DataFeeder object that returns training data.
Raises:
ValueError: if one of `x` and `y` is iterable and the other is not.
"""
x, y = _data_type_filter(x, y)
if HAS_DASK:
# pylint: disable=g-import-not-at-top
import dask.dataframe as dd
if (isinstance(x, (dd.Series, dd.DataFrame)) and
(y is None or isinstance(y, (dd.Series, dd.DataFrame)))):
data_feeder_cls = DaskDataFeeder
else:
data_feeder_cls = DataFeeder
else:
data_feeder_cls = DataFeeder
if _is_iterable(x):
if y is not None and not _is_iterable(y):
raise ValueError('Both x and y should be iterators for '
'streaming learning to work.')
return StreamingDataFeeder(x, y, n_classes, batch_size)
return data_feeder_cls(
x, y, n_classes, batch_size, shuffle=shuffle, epochs=epochs)
def _batch_data(x, batch_size=None):
if (batch_size is not None) and (batch_size <= 0):
raise ValueError('Invalid batch_size %d.' % batch_size)
x_first_el = six.next(x)
x = itertools.chain([x_first_el], x)
chunk = dict([(k, []) for k in list(x_first_el.keys())]) if isinstance(
x_first_el, dict) else []
chunk_filled = False
for data in x:
if isinstance(data, dict):
for k, v in list(data.items()):
chunk[k].append(v)
if (batch_size is not None) and (len(chunk[k]) >= batch_size):
chunk[k] = np.matrix(chunk[k])
chunk_filled = True
if chunk_filled:
yield chunk
chunk = dict([(k, []) for k in list(x_first_el.keys())]) if isinstance(
x_first_el, dict) else []
chunk_filled = False
else:
chunk.append(data)
if (batch_size is not None) and (len(chunk) >= batch_size):
yield np.matrix(chunk)
chunk = []
if isinstance(x_first_el, dict):
for k, v in list(data.items()):
chunk[k] = np.matrix(chunk[k])
yield chunk
else:
yield np.matrix(chunk)
def setup_predict_data_feeder(x, batch_size=None):
"""Returns an iterable for feeding into predict step.
Args:
x: numpy, pandas, Dask array or dictionary of aforementioned. Also supports
iterable.
batch_size: Size of batches to split data into. If `None`, returns one
batch of full size.
Returns:
List or iterator (or dictionary thereof) of parts of data to predict on.
Raises:
ValueError: if `batch_size` <= 0.
"""
if HAS_DASK:
x = extract_dask_data(x)
if HAS_PANDAS:
x = extract_pandas_data(x)
if _is_iterable(x):
return _batch_data(x, batch_size)
if len(x.shape) == 1:
x = np.reshape(x, (-1, 1))
if batch_size is not None:
if batch_size <= 0:
raise ValueError('Invalid batch_size %d.' % batch_size)
n_batches = int(math.ceil(float(len(x)) / batch_size))
return [x[i * batch_size:(i + 1) * batch_size] for i in xrange(n_batches)]
return [x]
def setup_processor_data_feeder(x):
"""Sets up processor iterable.
Args:
x: numpy, pandas or iterable.
Returns:
Iterable of data to process.
"""
if HAS_PANDAS:
x = extract_pandas_matrix(x)
return x
def check_array(array, dtype):
"""Checks array on dtype and converts it if different.
Args:
array: Input array.
dtype: Expected dtype.
Returns:
Original array or converted.
"""
# skip check if array is instance of other classes, e.g. h5py.Dataset
# to avoid copying array and loading whole data into memory
if isinstance(array, (np.ndarray, list)):
array = np.array(array, dtype=dtype, order=None, copy=False)
return array
def _access(data, iloc):
"""Accesses an element from collection, using integer location based indexing.
Args:
data: array-like. The collection to access
iloc: `int` or `list` of `int`s. Location(s) to access in `collection`
Returns:
The element of `a` found at location(s) `iloc`.
"""
if HAS_PANDAS:
import pandas as pd # pylint: disable=g-import-not-at-top
if isinstance(data, pd.Series) or isinstance(data, pd.DataFrame):
return data.iloc[iloc]
return data[iloc]
def _check_dtype(dtype):
if dtypes.as_dtype(dtype) == dtypes.float64:
logging.warn(
'float64 is not supported by many models, consider casting to float32.')
return dtype
class DataFeeder(object):
"""Data feeder is an example class to sample data for TF trainer."""
def __init__(self,
x,
y,
n_classes,
batch_size=None,
shuffle=True,
random_state=None,
epochs=None):
"""Initializes a DataFeeder instance.
Args:
x: One feature sample which can either Nd numpy matrix of shape
`[n_samples, n_features, ...]` or dictionary of Nd numpy matrix.
y: label vector, either floats for regression or class id for
classification. If matrix, will consider as a sequence of labels.
Can be `None` for unsupervised setting. Also supports dictionary of
labels.
n_classes: Number of classes, 0 and 1 are considered regression, `None`
will pass through the input labels without one-hot conversion. Also, if
`y` is `dict`, then `n_classes` must be `dict` such that
`n_classes[key] = n_classes for label y[key]`, `None` otherwise.
batch_size: Mini-batch size to accumulate samples in one mini batch.
shuffle: Whether to shuffle `x`.
random_state: Numpy `RandomState` object to reproduce sampling.
epochs: Number of times to iterate over input data before raising
`StopIteration` exception.
Attributes:
x: Input features (ndarray or dictionary of ndarrays).
y: Input label (ndarray or dictionary of ndarrays).
n_classes: Number of classes (if `None`, pass through indices without
one-hot conversion).
batch_size: Mini-batch size to accumulate.
input_shape: Shape of the input (or dictionary of shapes).
output_shape: Shape of the output (or dictionary of shapes).
input_dtype: DType of input (or dictionary of shapes).
output_dtype: DType of output (or dictionary of shapes.
"""
x_is_dict, y_is_dict = isinstance(x, dict), y is not None and isinstance(
y, dict)
if isinstance(y, list):
y = np.array(y)
self._x = dict([(k, check_array(v, v.dtype)) for k, v in list(x.items())
]) if x_is_dict else check_array(x, x.dtype)
self._y = None if y is None else \
dict([(k, check_array(v, v.dtype)) for k, v in list(y.items())]) if x_is_dict else check_array(y, y.dtype)
# self.n_classes is not None means we're converting raw target indices to one-hot.
if n_classes is not None:
if not y_is_dict:
y_dtype = (np.int64
if n_classes is not None and n_classes > 1 else np.float32)
self._y = (None if y is None else check_array(y, dtype=y_dtype))
self.n_classes = n_classes
self.max_epochs = epochs
x_shape = dict([(k, v.shape) for k, v in list(self._x.items())
]) if x_is_dict else self._x.shape
y_shape = dict([(k, v.shape) for k, v in list(self._y.items())
]) if y_is_dict else None if y is None else self._y.shape
self.input_shape, self.output_shape, self._batch_size = _get_in_out_shape(
x_shape, y_shape, n_classes, batch_size)
# Input dtype matches dtype of x.
self._input_dtype = dict([(k, _check_dtype(v.dtype)) for k, v in list(self._x.items())]) if x_is_dict \
else _check_dtype(self._x.dtype)
# note: self._output_dtype = np.float32 when y is None
self._output_dtype = dict([(k, _check_dtype(v.dtype)) for k, v in list(self._y.items())]) if y_is_dict \
else _check_dtype(self._y.dtype) if y is not None else np.float32
# self.n_classes is None means we're passing in raw target indices
if n_classes is not None and y_is_dict:
for key in list(n_classes.keys()):
if key in self._output_dtype:
self._output_dtype[key] = np.float32
self._shuffle = shuffle
self.random_state = np.random.RandomState(
42) if random_state is None else random_state
num_samples = list(self._x.values())[0].shape[
0] if x_is_dict else self._x.shape[0]
if self._shuffle:
self.indices = self.random_state.permutation(num_samples)
else:
self.indices = np.array(range(num_samples))
self.offset = 0
self.epoch = 0
self._epoch_placeholder = None
@property
def x(self):
return self._x
@property
def y(self):
return self._y
@property
def shuffle(self):
return self._shuffle
@property
def input_dtype(self):
return self._input_dtype
@property
def output_dtype(self):
return self._output_dtype
@property
def batch_size(self):
return self._batch_size
def make_epoch_variable(self):
"""Adds a placeholder variable for the epoch to the graph.
Returns:
The epoch placeholder.
"""
self._epoch_placeholder = array_ops.placeholder(
dtypes.int32, [1], name='epoch')
return self._epoch_placeholder
def input_builder(self):
"""Builds inputs in the graph.
Returns:
Two placeholders for inputs and outputs.
"""
def get_placeholder(shape, dtype, name_prepend):
if shape is None:
return None
if isinstance(shape, dict):
placeholder = {}
for key in list(shape.keys()):
placeholder[key] = array_ops.placeholder(
dtypes.as_dtype(dtype[key]), [None] + shape[key][1:],
name=name_prepend + '_' + key)
else:
placeholder = array_ops.placeholder(
dtypes.as_dtype(dtype), [None] + shape[1:], name=name_prepend)
return placeholder
self._input_placeholder = get_placeholder(self.input_shape,
self._input_dtype, 'input')
self._output_placeholder = get_placeholder(self.output_shape,
self._output_dtype, 'output')
return self._input_placeholder, self._output_placeholder
def set_placeholders(self, input_placeholder, output_placeholder):
"""Sets placeholders for this data feeder.
Args:
input_placeholder: Placeholder for `x` variable. Should match shape
of the examples in the x dataset.
output_placeholder: Placeholder for `y` variable. Should match
shape of the examples in the y dataset. Can be `None`.
"""
self._input_placeholder = input_placeholder
self._output_placeholder = output_placeholder
def get_feed_params(self):
"""Function returns a `dict` with data feed params while training.
Returns:
A `dict` with data feed params while training.
"""
return {
'epoch': self.epoch,
'offset': self.offset,
'batch_size': self._batch_size
}
def get_feed_dict_fn(self):
"""Returns a function that samples data into given placeholders.
Returns:
A function that when called samples a random subset of batch size
from `x` and `y`.
"""
x_is_dict, y_is_dict = isinstance(
self._x, dict), self._y is not None and isinstance(self._y, dict)
# Assign input features from random indices.
def extract(data, indices):
return (np.array(_access(data, indices)).reshape((indices.shape[0], 1)) if
len(data.shape) == 1 else _access(data, indices))
# assign labels from random indices
def assign_label(data, shape, dtype, n_classes, indices):
shape[0] = indices.shape[0]
out = np.zeros(shape, dtype=dtype)
for i in xrange(out.shape[0]):
sample = indices[i]
# self.n_classes is None means we're passing in raw target indices
if n_classes is None:
out[i] = _access(data, sample)
else:
if n_classes > 1:
if len(shape) == 2:
out.itemset((i, int(_access(data, sample))), 1.0)
else:
for idx, value in enumerate(_access(data, sample)):
out.itemset(tuple([i, idx, value]), 1.0)
else:
out[i] = _access(data, sample)
return out
def _feed_dict_fn():
"""Function that samples data into given placeholders."""
if self.max_epochs is not None and self.epoch + 1 > self.max_epochs:
raise StopIteration
assert self._input_placeholder is not None
feed_dict = {}
if self._epoch_placeholder is not None:
feed_dict[self._epoch_placeholder.name] = [self.epoch]
# Take next batch of indices.
x_len = list(self._x.values())[0].shape[
0] if x_is_dict else self._x.shape[0]
end = min(x_len, self.offset + self._batch_size)
batch_indices = self.indices[self.offset:end]
# adding input placeholder
feed_dict.update(
dict([(self._input_placeholder[k].name, extract(v, batch_indices))
for k, v in list(self._x.items())]) if x_is_dict else
{self._input_placeholder.name: extract(self._x, batch_indices)})
# move offset and reset it if necessary
self.offset += self._batch_size
if self.offset >= x_len:
self.indices = self.random_state.permutation(
x_len) if self._shuffle else np.array(range(x_len))
self.offset = 0
self.epoch += 1
# return early if there are no labels
if self._output_placeholder is None:
return feed_dict
# adding output placeholders
if y_is_dict:
for k, v in list(self._y.items()):
n_classes = (self.n_classes[k] if k in self.n_classes else
None) if self.n_classes is not None else None
shape, dtype = self.output_shape[k], self._output_dtype[k]
feed_dict.update({
self._output_placeholder[k].name:
assign_label(v, shape, dtype, n_classes, batch_indices)
})
else:
shape, dtype, n_classes = self.output_shape, self._output_dtype, self.n_classes
feed_dict.update({
self._output_placeholder.name:
assign_label(self._y, shape, dtype, n_classes, batch_indices)
})
return feed_dict
return _feed_dict_fn
class StreamingDataFeeder(DataFeeder):
"""Data feeder for TF trainer that reads data from iterator.
Streaming data feeder allows to read data as it comes it from disk or
somewhere else. It's custom to have this iterators rotate infinetly over
the dataset, to allow control of how much to learn on the trainer side.
"""
def __init__(self, x, y, n_classes, batch_size):
"""Initializes a StreamingDataFeeder instance.
Args:
x: iterator each element of which returns one feature sample. Sample can
be a Nd numpy matrix or dictionary of Nd numpy matrices.
y: iterator each element of which returns one label sample. Sample can be
a Nd numpy matrix or dictionary of Nd numpy matrices with 1 or many
classes regression values.
n_classes: indicator of how many classes the corresponding label sample
has for the purposes of one-hot conversion of label. In case where `y`
is a dictionary, `n_classes` must be dictionary (with same keys as `y`)
of how many classes there are in each label in `y`. If key is
present in `y` and missing in `n_classes`, the value is assumed `None`
and no one-hot conversion will be applied to the label with that key.
batch_size: Mini batch size to accumulate samples in one batch. If set
`None`, then assumes that iterator to return already batched element.
Attributes:
x: input features (or dictionary of input features).
y: input label (or dictionary of output features).
n_classes: number of classes.
batch_size: mini batch size to accumulate.
input_shape: shape of the input (can be dictionary depending on `x`).
output_shape: shape of the output (can be dictionary depending on `y`).
input_dtype: dtype of input (can be dictionary depending on `x`).
output_dtype: dtype of output (can be dictionary depending on `y`).
"""
# pylint: disable=invalid-name,super-init-not-called
x_first_el = six.next(x)
self._x = itertools.chain([x_first_el], x)
if y is not None:
y_first_el = six.next(y)
self._y = itertools.chain([y_first_el], y)
else:
y_first_el = None
self._y = None
self.n_classes = n_classes
x_is_dict = isinstance(x_first_el, dict)
y_is_dict = y is not None and isinstance(y_first_el, dict)
if y_is_dict and n_classes is not None:
assert isinstance(n_classes, dict)
# extract shapes for first_elements
if x_is_dict:
x_first_el_shape = dict(
[(k, [1] + list(v.shape)) for k, v in list(x_first_el.items())])
else:
x_first_el_shape = [1] + list(x_first_el.shape)
if y_is_dict:
y_first_el_shape = dict(
[(k, [1] + list(v.shape)) for k, v in list(y_first_el.items())])
elif y is None:
y_first_el_shape = None
else:
y_first_el_shape = ([1] + list(y_first_el[0].shape if isinstance(
y_first_el, list) else y_first_el.shape))
self.input_shape, self.output_shape, self._batch_size = _get_in_out_shape(
x_first_el_shape, y_first_el_shape, n_classes, batch_size)
# Input dtype of x_first_el.
if x_is_dict:
self._input_dtype = dict(
[(k, _check_dtype(v.dtype)) for k, v in list(x_first_el.items())])
else:
self._input_dtype = _check_dtype(x_first_el.dtype)
# Output dtype of y_first_el.
def check_y_dtype(el):
if isinstance(el, np.ndarray):
return el.dtype
elif isinstance(el, list):
return check_y_dtype(el[0])
else:
return _check_dtype(np.dtype(type(el)))
# Output types are floats, due to both softmaxes and regression req.
if n_classes is not None and (y is None or not y_is_dict) and n_classes > 0:
self._output_dtype = np.float32
elif y_is_dict:
self._output_dtype = dict(
[(k, check_y_dtype(v)) for k, v in list(y_first_el.items())])
elif y is None:
self._output_dtype = None
else:
self._output_dtype = check_y_dtype(y_first_el)
def get_feed_params(self):
"""Function returns a `dict` with data feed params while training.
Returns:
A `dict` with data feed params while training.
"""
return {'batch_size': self._batch_size}
def get_feed_dict_fn(self):
"""Returns a function, that will sample data and provide it to placeholders.
Returns:
A function that when called samples a random subset of batch size
from x and y.
"""
self.stopped = False
def _feed_dict_fn():
"""Samples data and provides it to placeholders.
Returns:
`dict` of input and output tensors.
"""
def init_array(shape, dtype):
"""Initialize array of given shape or dict of shapes and dtype."""
if shape is None:
return None
elif isinstance(shape, dict):
return dict([(k, np.zeros(shape[k], dtype[k]))
for k in list(shape.keys())])
else:
return np.zeros(shape, dtype=dtype)
def put_data_array(dest, index, source=None, n_classes=None):
"""Puts data array into container."""
if source is None:
dest = dest[:index]
elif n_classes is not None and n_classes > 1:
if len(self.output_shape) == 2:
dest.itemset((index, source), 1.0)
else:
for idx, value in enumerate(source):
dest.itemset(tuple([index, idx, value]), 1.0)
else:
if len(dest.shape) > 1:
dest[index, :] = source
else:
dest[index] = source[0] if isinstance(source, list) else source
return dest
def put_data_array_or_dict(holder, index, data=None, n_classes=None):
"""Puts data array or data dictionary into container."""
if holder is None:
return None
if isinstance(holder, dict):
if data is None:
data = {k: None for k in holder.keys()}
assert isinstance(data, dict)
for k in holder.keys():
num_classes = n_classes[k] if (n_classes is not None and
k in n_classes) else None
holder[k] = put_data_array(holder[k], index, data[k], num_classes)
else:
holder = put_data_array(holder, index, data, n_classes)
return holder
if self.stopped:
raise StopIteration
inp = init_array(self.input_shape, self._input_dtype)
out = init_array(self.output_shape, self._output_dtype)
for i in xrange(self._batch_size):
# Add handling when queue ends.
try:
next_inp = six.next(self._x)
inp = put_data_array_or_dict(inp, i, next_inp, None)
except StopIteration:
self.stopped = True
if i == 0:
raise
inp = put_data_array_or_dict(inp, i, None, None)
out = put_data_array_or_dict(out, i, None, None)
break
if self._y is not None:
next_out = six.next(self._y)
out = put_data_array_or_dict(out, i, next_out, self.n_classes)
# creating feed_dict
if isinstance(inp, dict):
feed_dict = dict([(self._input_placeholder[k].name, inp[k])
for k in list(self._input_placeholder.keys())])
else:
feed_dict = {self._input_placeholder.name: inp}
if self._y is not None:
if isinstance(out, dict):
feed_dict.update(
dict([(self._output_placeholder[k].name, out[k])
for k in list(self._output_placeholder.keys())]))
else:
feed_dict.update({self._output_placeholder.name: out})
return feed_dict
return _feed_dict_fn
class DaskDataFeeder(object):
"""Data feeder for that reads data from dask.Series and dask.DataFrame.
Numpy arrays can be serialized to disk and it's possible to do random seeks
into them. DaskDataFeeder will remove requirement to have full dataset in the
memory and still do random seeks for sampling of batches.
"""
def __init__(self,
x,
y,
n_classes,
batch_size,
shuffle=True,
random_state=None,
epochs=None):
"""Initializes a DaskDataFeeder instance.
Args:
x: iterator that returns for each element, returns features.
y: iterator that returns for each element, returns 1 or many classes /
regression values.
n_classes: indicator of how many classes the label has.
batch_size: Mini batch size to accumulate.
shuffle: Whether to shuffle the inputs.
random_state: random state for RNG. Note that it will mutate so use a
int value for this if you want consistent sized batches.
epochs: Number of epochs to run.
Attributes:
x: input features.
y: input label.
n_classes: number of classes.
batch_size: mini batch size to accumulate.
input_shape: shape of the input.
output_shape: shape of the output.
input_dtype: dtype of input.
output_dtype: dtype of output.
Raises:
ValueError: if `x` or `y` are `dict`, as they are not supported currently.
"""
if isinstance(x, dict) or isinstance(y, dict):
raise ValueError(
'DaskDataFeeder does not support dictionaries at the moment.')
# pylint: disable=invalid-name,super-init-not-called
import dask.dataframe as dd # pylint: disable=g-import-not-at-top
# TODO(terrytangyuan): check x and y dtypes in dask_io like pandas
self._x = x
self._y = y
# save column names
self._x_columns = list(x.columns)
if isinstance(y.columns[0], str):
self._y_columns = list(y.columns)
else:
# deal with cases where two DFs have overlapped default numeric colnames
self._y_columns = len(self._x_columns) + 1
self._y = self._y.rename(columns={y.columns[0]: self._y_columns})
# TODO(terrytangyuan): deal with unsupervised cases
# combine into a data frame
self.df = dd.multi.concat([self._x, self._y], axis=1)
self.n_classes = n_classes
x_count = x.count().compute()[0]
x_shape = (x_count, len(self._x.columns))
y_shape = (x_count, len(self._y.columns))
# TODO(terrytangyuan): Add support for shuffle and epochs.
self._shuffle = shuffle
self.epochs = epochs
self.input_shape, self.output_shape, self._batch_size = _get_in_out_shape(
x_shape, y_shape, n_classes, batch_size)
self.sample_fraction = self._batch_size / float(x_count)
self._input_dtype = _check_dtype(self._x.dtypes[0])
self._output_dtype = _check_dtype(self._y.dtypes[self._y_columns])
if random_state is None:
self.random_state = 66
else:
self.random_state = random_state
def get_feed_params(self):
"""Function returns a `dict` with data feed params while training.
Returns:
A `dict` with data feed params while training.
"""
return {'batch_size': self._batch_size}
def get_feed_dict_fn(self, input_placeholder, output_placeholder):
"""Returns a function, that will sample data and provide it to placeholders.
Args:
input_placeholder: tf.Placeholder for input features mini batch.
output_placeholder: tf.Placeholder for output labels.
Returns:
A function that when called samples a random subset of batch size
from x and y.
"""
def _feed_dict_fn():
"""Samples data and provides it to placeholders."""
# TODO(ipolosukhin): option for with/without replacement (dev version of
# dask)
sample = self.df.random_split(
[self.sample_fraction, 1 - self.sample_fraction],
random_state=self.random_state)
inp = extract_pandas_matrix(sample[0][self._x_columns].compute()).tolist()
out = extract_pandas_matrix(sample[0][self._y_columns].compute())
# convert to correct dtype
inp = np.array(inp, dtype=self._input_dtype)
# one-hot encode out for each class for cross entropy loss
if HAS_PANDAS:
import pandas as pd # pylint: disable=g-import-not-at-top
if not isinstance(out, pd.Series):
out = out.flatten()
out_max = self._y.max().compute().values[0]
encoded_out = np.zeros((out.size, out_max + 1), dtype=self._output_dtype)
encoded_out[np.arange(out.size), out] = 1
return {input_placeholder.name: inp, output_placeholder.name: encoded_out}
return _feed_dict_fn
| apache-2.0 |
eg-zhang/scikit-learn | sklearn/ensemble/weight_boosting.py | 71 | 40664 | """Weight Boosting
This module contains weight boosting estimators for both classification and
regression.
The module structure is the following:
- The ``BaseWeightBoosting`` base class implements a common ``fit`` method
for all the estimators in the module. Regression and classification
only differ from each other in the loss function that is optimized.
- ``AdaBoostClassifier`` implements adaptive boosting (AdaBoost-SAMME) for
classification problems.
- ``AdaBoostRegressor`` implements adaptive boosting (AdaBoost.R2) for
regression problems.
"""
# Authors: Noel Dawe <noel@dawe.me>
# Gilles Louppe <g.louppe@gmail.com>
# Hamzeh Alsalhi <ha258@cornell.edu>
# Arnaud Joly <arnaud.v.joly@gmail.com>
#
# Licence: BSD 3 clause
from abc import ABCMeta, abstractmethod
import numpy as np
from numpy.core.umath_tests import inner1d
from .base import BaseEnsemble
from ..base import ClassifierMixin, RegressorMixin
from ..externals import six
from ..externals.six.moves import zip
from ..externals.six.moves import xrange as range
from .forest import BaseForest
from ..tree import DecisionTreeClassifier, DecisionTreeRegressor
from ..tree.tree import BaseDecisionTree
from ..tree._tree import DTYPE
from ..utils import check_array, check_X_y, check_random_state
from ..metrics import accuracy_score, r2_score
from sklearn.utils.validation import has_fit_parameter, check_is_fitted
__all__ = [
'AdaBoostClassifier',
'AdaBoostRegressor',
]
class BaseWeightBoosting(six.with_metaclass(ABCMeta, BaseEnsemble)):
"""Base class for AdaBoost estimators.
Warning: This class should not be used directly. Use derived classes
instead.
"""
@abstractmethod
def __init__(self,
base_estimator=None,
n_estimators=50,
estimator_params=tuple(),
learning_rate=1.,
random_state=None):
super(BaseWeightBoosting, self).__init__(
base_estimator=base_estimator,
n_estimators=n_estimators,
estimator_params=estimator_params)
self.learning_rate = learning_rate
self.random_state = random_state
def fit(self, X, y, sample_weight=None):
"""Build a boosted classifier/regressor from the training set (X, y).
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. COO, DOK, and LIL are converted to CSR. The dtype is
forced to DTYPE from tree._tree if the base classifier of this
ensemble weighted boosting classifier is a tree or forest.
y : array-like of shape = [n_samples]
The target values (class labels in classification, real numbers in
regression).
sample_weight : array-like of shape = [n_samples], optional
Sample weights. If None, the sample weights are initialized to
1 / n_samples.
Returns
-------
self : object
Returns self.
"""
# Check parameters
if self.learning_rate <= 0:
raise ValueError("learning_rate must be greater than zero")
if (self.base_estimator is None or
isinstance(self.base_estimator, (BaseDecisionTree,
BaseForest))):
dtype = DTYPE
accept_sparse = 'csc'
else:
dtype = None
accept_sparse = ['csr', 'csc']
X, y = check_X_y(X, y, accept_sparse=accept_sparse, dtype=dtype)
if sample_weight is None:
# Initialize weights to 1 / n_samples
sample_weight = np.empty(X.shape[0], dtype=np.float)
sample_weight[:] = 1. / X.shape[0]
else:
# Normalize existing weights
sample_weight = sample_weight / sample_weight.sum(dtype=np.float64)
# Check that the sample weights sum is positive
if sample_weight.sum() <= 0:
raise ValueError(
"Attempting to fit with a non-positive "
"weighted number of samples.")
# Check parameters
self._validate_estimator()
# Clear any previous fit results
self.estimators_ = []
self.estimator_weights_ = np.zeros(self.n_estimators, dtype=np.float)
self.estimator_errors_ = np.ones(self.n_estimators, dtype=np.float)
for iboost in range(self.n_estimators):
# Boosting step
sample_weight, estimator_weight, estimator_error = self._boost(
iboost,
X, y,
sample_weight)
# Early termination
if sample_weight is None:
break
self.estimator_weights_[iboost] = estimator_weight
self.estimator_errors_[iboost] = estimator_error
# Stop if error is zero
if estimator_error == 0:
break
sample_weight_sum = np.sum(sample_weight)
# Stop if the sum of sample weights has become non-positive
if sample_weight_sum <= 0:
break
if iboost < self.n_estimators - 1:
# Normalize
sample_weight /= sample_weight_sum
return self
@abstractmethod
def _boost(self, iboost, X, y, sample_weight):
"""Implement a single boost.
Warning: This method needs to be overriden by subclasses.
Parameters
----------
iboost : int
The index of the current boost iteration.
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. COO, DOK, and LIL are converted to CSR.
y : array-like of shape = [n_samples]
The target values (class labels).
sample_weight : array-like of shape = [n_samples]
The current sample weights.
Returns
-------
sample_weight : array-like of shape = [n_samples] or None
The reweighted sample weights.
If None then boosting has terminated early.
estimator_weight : float
The weight for the current boost.
If None then boosting has terminated early.
error : float
The classification error for the current boost.
If None then boosting has terminated early.
"""
pass
def staged_score(self, X, y, sample_weight=None):
"""Return staged scores for X, y.
This generator method yields the ensemble score after each iteration of
boosting and therefore allows monitoring, such as to determine the
score on a test set after each boost.
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
y : array-like, shape = [n_samples]
Labels for X.
sample_weight : array-like, shape = [n_samples], optional
Sample weights.
Returns
-------
z : float
"""
for y_pred in self.staged_predict(X):
if isinstance(self, ClassifierMixin):
yield accuracy_score(y, y_pred, sample_weight=sample_weight)
else:
yield r2_score(y, y_pred, sample_weight=sample_weight)
@property
def feature_importances_(self):
"""Return the feature importances (the higher, the more important the
feature).
Returns
-------
feature_importances_ : array, shape = [n_features]
"""
if self.estimators_ is None or len(self.estimators_) == 0:
raise ValueError("Estimator not fitted, "
"call `fit` before `feature_importances_`.")
try:
norm = self.estimator_weights_.sum()
return (sum(weight * clf.feature_importances_ for weight, clf
in zip(self.estimator_weights_, self.estimators_))
/ norm)
except AttributeError:
raise AttributeError(
"Unable to compute feature importances "
"since base_estimator does not have a "
"feature_importances_ attribute")
def _validate_X_predict(self, X):
"""Ensure that X is in the proper format"""
if (self.base_estimator is None or
isinstance(self.base_estimator,
(BaseDecisionTree, BaseForest))):
X = check_array(X, accept_sparse='csr', dtype=DTYPE)
else:
X = check_array(X, accept_sparse=['csr', 'csc', 'coo'])
return X
def _samme_proba(estimator, n_classes, X):
"""Calculate algorithm 4, step 2, equation c) of Zhu et al [1].
References
----------
.. [1] J. Zhu, H. Zou, S. Rosset, T. Hastie, "Multi-class AdaBoost", 2009.
"""
proba = estimator.predict_proba(X)
# Displace zero probabilities so the log is defined.
# Also fix negative elements which may occur with
# negative sample weights.
proba[proba < np.finfo(proba.dtype).eps] = np.finfo(proba.dtype).eps
log_proba = np.log(proba)
return (n_classes - 1) * (log_proba - (1. / n_classes)
* log_proba.sum(axis=1)[:, np.newaxis])
class AdaBoostClassifier(BaseWeightBoosting, ClassifierMixin):
"""An AdaBoost classifier.
An AdaBoost [1] classifier is a meta-estimator that begins by fitting a
classifier on the original dataset and then fits additional copies of the
classifier on the same dataset but where the weights of incorrectly
classified instances are adjusted such that subsequent classifiers focus
more on difficult cases.
This class implements the algorithm known as AdaBoost-SAMME [2].
Read more in the :ref:`User Guide <adaboost>`.
Parameters
----------
base_estimator : object, optional (default=DecisionTreeClassifier)
The base estimator from which the boosted ensemble is built.
Support for sample weighting is required, as well as proper `classes_`
and `n_classes_` attributes.
n_estimators : integer, optional (default=50)
The maximum number of estimators at which boosting is terminated.
In case of perfect fit, the learning procedure is stopped early.
learning_rate : float, optional (default=1.)
Learning rate shrinks the contribution of each classifier by
``learning_rate``. There is a trade-off between ``learning_rate`` and
``n_estimators``.
algorithm : {'SAMME', 'SAMME.R'}, optional (default='SAMME.R')
If 'SAMME.R' then use the SAMME.R real boosting algorithm.
``base_estimator`` must support calculation of class probabilities.
If 'SAMME' then use the SAMME discrete boosting algorithm.
The SAMME.R algorithm typically converges faster than SAMME,
achieving a lower test error with fewer boosting iterations.
random_state : int, RandomState instance or None, optional (default=None)
If int, random_state is the seed used by the random number generator;
If RandomState instance, random_state is the random number generator;
If None, the random number generator is the RandomState instance used
by `np.random`.
Attributes
----------
estimators_ : list of classifiers
The collection of fitted sub-estimators.
classes_ : array of shape = [n_classes]
The classes labels.
n_classes_ : int
The number of classes.
estimator_weights_ : array of floats
Weights for each estimator in the boosted ensemble.
estimator_errors_ : array of floats
Classification error for each estimator in the boosted
ensemble.
feature_importances_ : array of shape = [n_features]
The feature importances if supported by the ``base_estimator``.
See also
--------
AdaBoostRegressor, GradientBoostingClassifier, DecisionTreeClassifier
References
----------
.. [1] Y. Freund, R. Schapire, "A Decision-Theoretic Generalization of
on-Line Learning and an Application to Boosting", 1995.
.. [2] J. Zhu, H. Zou, S. Rosset, T. Hastie, "Multi-class AdaBoost", 2009.
"""
def __init__(self,
base_estimator=None,
n_estimators=50,
learning_rate=1.,
algorithm='SAMME.R',
random_state=None):
super(AdaBoostClassifier, self).__init__(
base_estimator=base_estimator,
n_estimators=n_estimators,
learning_rate=learning_rate,
random_state=random_state)
self.algorithm = algorithm
def fit(self, X, y, sample_weight=None):
"""Build a boosted classifier from the training set (X, y).
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
y : array-like of shape = [n_samples]
The target values (class labels).
sample_weight : array-like of shape = [n_samples], optional
Sample weights. If None, the sample weights are initialized to
``1 / n_samples``.
Returns
-------
self : object
Returns self.
"""
# Check that algorithm is supported
if self.algorithm not in ('SAMME', 'SAMME.R'):
raise ValueError("algorithm %s is not supported" % self.algorithm)
# Fit
return super(AdaBoostClassifier, self).fit(X, y, sample_weight)
def _validate_estimator(self):
"""Check the estimator and set the base_estimator_ attribute."""
super(AdaBoostClassifier, self)._validate_estimator(
default=DecisionTreeClassifier(max_depth=1))
# SAMME-R requires predict_proba-enabled base estimators
if self.algorithm == 'SAMME.R':
if not hasattr(self.base_estimator_, 'predict_proba'):
raise TypeError(
"AdaBoostClassifier with algorithm='SAMME.R' requires "
"that the weak learner supports the calculation of class "
"probabilities with a predict_proba method.\n"
"Please change the base estimator or set "
"algorithm='SAMME' instead.")
if not has_fit_parameter(self.base_estimator_, "sample_weight"):
raise ValueError("%s doesn't support sample_weight."
% self.base_estimator_.__class__.__name__)
def _boost(self, iboost, X, y, sample_weight):
"""Implement a single boost.
Perform a single boost according to the real multi-class SAMME.R
algorithm or to the discrete SAMME algorithm and return the updated
sample weights.
Parameters
----------
iboost : int
The index of the current boost iteration.
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
y : array-like of shape = [n_samples]
The target values (class labels).
sample_weight : array-like of shape = [n_samples]
The current sample weights.
Returns
-------
sample_weight : array-like of shape = [n_samples] or None
The reweighted sample weights.
If None then boosting has terminated early.
estimator_weight : float
The weight for the current boost.
If None then boosting has terminated early.
estimator_error : float
The classification error for the current boost.
If None then boosting has terminated early.
"""
if self.algorithm == 'SAMME.R':
return self._boost_real(iboost, X, y, sample_weight)
else: # elif self.algorithm == "SAMME":
return self._boost_discrete(iboost, X, y, sample_weight)
def _boost_real(self, iboost, X, y, sample_weight):
"""Implement a single boost using the SAMME.R real algorithm."""
estimator = self._make_estimator()
try:
estimator.set_params(random_state=self.random_state)
except ValueError:
pass
estimator.fit(X, y, sample_weight=sample_weight)
y_predict_proba = estimator.predict_proba(X)
if iboost == 0:
self.classes_ = getattr(estimator, 'classes_', None)
self.n_classes_ = len(self.classes_)
y_predict = self.classes_.take(np.argmax(y_predict_proba, axis=1),
axis=0)
# Instances incorrectly classified
incorrect = y_predict != y
# Error fraction
estimator_error = np.mean(
np.average(incorrect, weights=sample_weight, axis=0))
# Stop if classification is perfect
if estimator_error <= 0:
return sample_weight, 1., 0.
# Construct y coding as described in Zhu et al [2]:
#
# y_k = 1 if c == k else -1 / (K - 1)
#
# where K == n_classes_ and c, k in [0, K) are indices along the second
# axis of the y coding with c being the index corresponding to the true
# class label.
n_classes = self.n_classes_
classes = self.classes_
y_codes = np.array([-1. / (n_classes - 1), 1.])
y_coding = y_codes.take(classes == y[:, np.newaxis])
# Displace zero probabilities so the log is defined.
# Also fix negative elements which may occur with
# negative sample weights.
proba = y_predict_proba # alias for readability
proba[proba < np.finfo(proba.dtype).eps] = np.finfo(proba.dtype).eps
# Boost weight using multi-class AdaBoost SAMME.R alg
estimator_weight = (-1. * self.learning_rate
* (((n_classes - 1.) / n_classes) *
inner1d(y_coding, np.log(y_predict_proba))))
# Only boost the weights if it will fit again
if not iboost == self.n_estimators - 1:
# Only boost positive weights
sample_weight *= np.exp(estimator_weight *
((sample_weight > 0) |
(estimator_weight < 0)))
return sample_weight, 1., estimator_error
def _boost_discrete(self, iboost, X, y, sample_weight):
"""Implement a single boost using the SAMME discrete algorithm."""
estimator = self._make_estimator()
try:
estimator.set_params(random_state=self.random_state)
except ValueError:
pass
estimator.fit(X, y, sample_weight=sample_weight)
y_predict = estimator.predict(X)
if iboost == 0:
self.classes_ = getattr(estimator, 'classes_', None)
self.n_classes_ = len(self.classes_)
# Instances incorrectly classified
incorrect = y_predict != y
# Error fraction
estimator_error = np.mean(
np.average(incorrect, weights=sample_weight, axis=0))
# Stop if classification is perfect
if estimator_error <= 0:
return sample_weight, 1., 0.
n_classes = self.n_classes_
# Stop if the error is at least as bad as random guessing
if estimator_error >= 1. - (1. / n_classes):
self.estimators_.pop(-1)
if len(self.estimators_) == 0:
raise ValueError('BaseClassifier in AdaBoostClassifier '
'ensemble is worse than random, ensemble '
'can not be fit.')
return None, None, None
# Boost weight using multi-class AdaBoost SAMME alg
estimator_weight = self.learning_rate * (
np.log((1. - estimator_error) / estimator_error) +
np.log(n_classes - 1.))
# Only boost the weights if I will fit again
if not iboost == self.n_estimators - 1:
# Only boost positive weights
sample_weight *= np.exp(estimator_weight * incorrect *
((sample_weight > 0) |
(estimator_weight < 0)))
return sample_weight, estimator_weight, estimator_error
def predict(self, X):
"""Predict classes for X.
The predicted class of an input sample is computed as the weighted mean
prediction of the classifiers in the ensemble.
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
Returns
-------
y : array of shape = [n_samples]
The predicted classes.
"""
pred = self.decision_function(X)
if self.n_classes_ == 2:
return self.classes_.take(pred > 0, axis=0)
return self.classes_.take(np.argmax(pred, axis=1), axis=0)
def staged_predict(self, X):
"""Return staged predictions for X.
The predicted class of an input sample is computed as the weighted mean
prediction of the classifiers in the ensemble.
This generator method yields the ensemble prediction after each
iteration of boosting and therefore allows monitoring, such as to
determine the prediction on a test set after each boost.
Parameters
----------
X : array-like of shape = [n_samples, n_features]
The input samples.
Returns
-------
y : generator of array, shape = [n_samples]
The predicted classes.
"""
n_classes = self.n_classes_
classes = self.classes_
if n_classes == 2:
for pred in self.staged_decision_function(X):
yield np.array(classes.take(pred > 0, axis=0))
else:
for pred in self.staged_decision_function(X):
yield np.array(classes.take(
np.argmax(pred, axis=1), axis=0))
def decision_function(self, X):
"""Compute the decision function of ``X``.
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
Returns
-------
score : array, shape = [n_samples, k]
The decision function of the input samples. The order of
outputs is the same of that of the `classes_` attribute.
Binary classification is a special cases with ``k == 1``,
otherwise ``k==n_classes``. For binary classification,
values closer to -1 or 1 mean more like the first or second
class in ``classes_``, respectively.
"""
check_is_fitted(self, "n_classes_")
X = self._validate_X_predict(X)
n_classes = self.n_classes_
classes = self.classes_[:, np.newaxis]
pred = None
if self.algorithm == 'SAMME.R':
# The weights are all 1. for SAMME.R
pred = sum(_samme_proba(estimator, n_classes, X)
for estimator in self.estimators_)
else: # self.algorithm == "SAMME"
pred = sum((estimator.predict(X) == classes).T * w
for estimator, w in zip(self.estimators_,
self.estimator_weights_))
pred /= self.estimator_weights_.sum()
if n_classes == 2:
pred[:, 0] *= -1
return pred.sum(axis=1)
return pred
def staged_decision_function(self, X):
"""Compute decision function of ``X`` for each boosting iteration.
This method allows monitoring (i.e. determine error on testing set)
after each boosting iteration.
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
Returns
-------
score : generator of array, shape = [n_samples, k]
The decision function of the input samples. The order of
outputs is the same of that of the `classes_` attribute.
Binary classification is a special cases with ``k == 1``,
otherwise ``k==n_classes``. For binary classification,
values closer to -1 or 1 mean more like the first or second
class in ``classes_``, respectively.
"""
check_is_fitted(self, "n_classes_")
X = self._validate_X_predict(X)
n_classes = self.n_classes_
classes = self.classes_[:, np.newaxis]
pred = None
norm = 0.
for weight, estimator in zip(self.estimator_weights_,
self.estimators_):
norm += weight
if self.algorithm == 'SAMME.R':
# The weights are all 1. for SAMME.R
current_pred = _samme_proba(estimator, n_classes, X)
else: # elif self.algorithm == "SAMME":
current_pred = estimator.predict(X)
current_pred = (current_pred == classes).T * weight
if pred is None:
pred = current_pred
else:
pred += current_pred
if n_classes == 2:
tmp_pred = np.copy(pred)
tmp_pred[:, 0] *= -1
yield (tmp_pred / norm).sum(axis=1)
else:
yield pred / norm
def predict_proba(self, X):
"""Predict class probabilities for X.
The predicted class probabilities of an input sample is computed as
the weighted mean predicted class probabilities of the classifiers
in the ensemble.
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
Returns
-------
p : array of shape = [n_samples]
The class probabilities of the input samples. The order of
outputs is the same of that of the `classes_` attribute.
"""
check_is_fitted(self, "n_classes_")
n_classes = self.n_classes_
X = self._validate_X_predict(X)
if self.algorithm == 'SAMME.R':
# The weights are all 1. for SAMME.R
proba = sum(_samme_proba(estimator, n_classes, X)
for estimator in self.estimators_)
else: # self.algorithm == "SAMME"
proba = sum(estimator.predict_proba(X) * w
for estimator, w in zip(self.estimators_,
self.estimator_weights_))
proba /= self.estimator_weights_.sum()
proba = np.exp((1. / (n_classes - 1)) * proba)
normalizer = proba.sum(axis=1)[:, np.newaxis]
normalizer[normalizer == 0.0] = 1.0
proba /= normalizer
return proba
def staged_predict_proba(self, X):
"""Predict class probabilities for X.
The predicted class probabilities of an input sample is computed as
the weighted mean predicted class probabilities of the classifiers
in the ensemble.
This generator method yields the ensemble predicted class probabilities
after each iteration of boosting and therefore allows monitoring, such
as to determine the predicted class probabilities on a test set after
each boost.
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
Returns
-------
p : generator of array, shape = [n_samples]
The class probabilities of the input samples. The order of
outputs is the same of that of the `classes_` attribute.
"""
X = self._validate_X_predict(X)
n_classes = self.n_classes_
proba = None
norm = 0.
for weight, estimator in zip(self.estimator_weights_,
self.estimators_):
norm += weight
if self.algorithm == 'SAMME.R':
# The weights are all 1. for SAMME.R
current_proba = _samme_proba(estimator, n_classes, X)
else: # elif self.algorithm == "SAMME":
current_proba = estimator.predict_proba(X) * weight
if proba is None:
proba = current_proba
else:
proba += current_proba
real_proba = np.exp((1. / (n_classes - 1)) * (proba / norm))
normalizer = real_proba.sum(axis=1)[:, np.newaxis]
normalizer[normalizer == 0.0] = 1.0
real_proba /= normalizer
yield real_proba
def predict_log_proba(self, X):
"""Predict class log-probabilities for X.
The predicted class log-probabilities of an input sample is computed as
the weighted mean predicted class log-probabilities of the classifiers
in the ensemble.
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
Returns
-------
p : array of shape = [n_samples]
The class probabilities of the input samples. The order of
outputs is the same of that of the `classes_` attribute.
"""
return np.log(self.predict_proba(X))
class AdaBoostRegressor(BaseWeightBoosting, RegressorMixin):
"""An AdaBoost regressor.
An AdaBoost [1] regressor is a meta-estimator that begins by fitting a
regressor on the original dataset and then fits additional copies of the
regressor on the same dataset but where the weights of instances are
adjusted according to the error of the current prediction. As such,
subsequent regressors focus more on difficult cases.
This class implements the algorithm known as AdaBoost.R2 [2].
Read more in the :ref:`User Guide <adaboost>`.
Parameters
----------
base_estimator : object, optional (default=DecisionTreeRegressor)
The base estimator from which the boosted ensemble is built.
Support for sample weighting is required.
n_estimators : integer, optional (default=50)
The maximum number of estimators at which boosting is terminated.
In case of perfect fit, the learning procedure is stopped early.
learning_rate : float, optional (default=1.)
Learning rate shrinks the contribution of each regressor by
``learning_rate``. There is a trade-off between ``learning_rate`` and
``n_estimators``.
loss : {'linear', 'square', 'exponential'}, optional (default='linear')
The loss function to use when updating the weights after each
boosting iteration.
random_state : int, RandomState instance or None, optional (default=None)
If int, random_state is the seed used by the random number generator;
If RandomState instance, random_state is the random number generator;
If None, the random number generator is the RandomState instance used
by `np.random`.
Attributes
----------
estimators_ : list of classifiers
The collection of fitted sub-estimators.
estimator_weights_ : array of floats
Weights for each estimator in the boosted ensemble.
estimator_errors_ : array of floats
Regression error for each estimator in the boosted ensemble.
feature_importances_ : array of shape = [n_features]
The feature importances if supported by the ``base_estimator``.
See also
--------
AdaBoostClassifier, GradientBoostingRegressor, DecisionTreeRegressor
References
----------
.. [1] Y. Freund, R. Schapire, "A Decision-Theoretic Generalization of
on-Line Learning and an Application to Boosting", 1995.
.. [2] H. Drucker, "Improving Regressors using Boosting Techniques", 1997.
"""
def __init__(self,
base_estimator=None,
n_estimators=50,
learning_rate=1.,
loss='linear',
random_state=None):
super(AdaBoostRegressor, self).__init__(
base_estimator=base_estimator,
n_estimators=n_estimators,
learning_rate=learning_rate,
random_state=random_state)
self.loss = loss
self.random_state = random_state
def fit(self, X, y, sample_weight=None):
"""Build a boosted regressor from the training set (X, y).
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
y : array-like of shape = [n_samples]
The target values (real numbers).
sample_weight : array-like of shape = [n_samples], optional
Sample weights. If None, the sample weights are initialized to
1 / n_samples.
Returns
-------
self : object
Returns self.
"""
# Check loss
if self.loss not in ('linear', 'square', 'exponential'):
raise ValueError(
"loss must be 'linear', 'square', or 'exponential'")
# Fit
return super(AdaBoostRegressor, self).fit(X, y, sample_weight)
def _validate_estimator(self):
"""Check the estimator and set the base_estimator_ attribute."""
super(AdaBoostRegressor, self)._validate_estimator(
default=DecisionTreeRegressor(max_depth=3))
def _boost(self, iboost, X, y, sample_weight):
"""Implement a single boost for regression
Perform a single boost according to the AdaBoost.R2 algorithm and
return the updated sample weights.
Parameters
----------
iboost : int
The index of the current boost iteration.
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
y : array-like of shape = [n_samples]
The target values (class labels in classification, real numbers in
regression).
sample_weight : array-like of shape = [n_samples]
The current sample weights.
Returns
-------
sample_weight : array-like of shape = [n_samples] or None
The reweighted sample weights.
If None then boosting has terminated early.
estimator_weight : float
The weight for the current boost.
If None then boosting has terminated early.
estimator_error : float
The regression error for the current boost.
If None then boosting has terminated early.
"""
estimator = self._make_estimator()
try:
estimator.set_params(random_state=self.random_state)
except ValueError:
pass
generator = check_random_state(self.random_state)
# Weighted sampling of the training set with replacement
# For NumPy >= 1.7.0 use np.random.choice
cdf = sample_weight.cumsum()
cdf /= cdf[-1]
uniform_samples = generator.random_sample(X.shape[0])
bootstrap_idx = cdf.searchsorted(uniform_samples, side='right')
# searchsorted returns a scalar
bootstrap_idx = np.array(bootstrap_idx, copy=False)
# Fit on the bootstrapped sample and obtain a prediction
# for all samples in the training set
estimator.fit(X[bootstrap_idx], y[bootstrap_idx])
y_predict = estimator.predict(X)
error_vect = np.abs(y_predict - y)
error_max = error_vect.max()
if error_max != 0.:
error_vect /= error_max
if self.loss == 'square':
error_vect **= 2
elif self.loss == 'exponential':
error_vect = 1. - np.exp(- error_vect)
# Calculate the average loss
estimator_error = (sample_weight * error_vect).sum()
if estimator_error <= 0:
# Stop if fit is perfect
return sample_weight, 1., 0.
elif estimator_error >= 0.5:
# Discard current estimator only if it isn't the only one
if len(self.estimators_) > 1:
self.estimators_.pop(-1)
return None, None, None
beta = estimator_error / (1. - estimator_error)
# Boost weight using AdaBoost.R2 alg
estimator_weight = self.learning_rate * np.log(1. / beta)
if not iboost == self.n_estimators - 1:
sample_weight *= np.power(
beta,
(1. - error_vect) * self.learning_rate)
return sample_weight, estimator_weight, estimator_error
def _get_median_predict(self, X, limit):
# Evaluate predictions of all estimators
predictions = np.array([
est.predict(X) for est in self.estimators_[:limit]]).T
# Sort the predictions
sorted_idx = np.argsort(predictions, axis=1)
# Find index of median prediction for each sample
weight_cdf = self.estimator_weights_[sorted_idx].cumsum(axis=1)
median_or_above = weight_cdf >= 0.5 * weight_cdf[:, -1][:, np.newaxis]
median_idx = median_or_above.argmax(axis=1)
median_estimators = sorted_idx[np.arange(X.shape[0]), median_idx]
# Return median predictions
return predictions[np.arange(X.shape[0]), median_estimators]
def predict(self, X):
"""Predict regression value for X.
The predicted regression value of an input sample is computed
as the weighted median prediction of the classifiers in the ensemble.
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
Returns
-------
y : array of shape = [n_samples]
The predicted regression values.
"""
check_is_fitted(self, "estimator_weights_")
X = self._validate_X_predict(X)
return self._get_median_predict(X, len(self.estimators_))
def staged_predict(self, X):
"""Return staged predictions for X.
The predicted regression value of an input sample is computed
as the weighted median prediction of the classifiers in the ensemble.
This generator method yields the ensemble prediction after each
iteration of boosting and therefore allows monitoring, such as to
determine the prediction on a test set after each boost.
Parameters
----------
X : {array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrix can be CSC, CSR, COO,
DOK, or LIL. DOK and LIL are converted to CSR.
Returns
-------
y : generator of array, shape = [n_samples]
The predicted regression values.
"""
check_is_fitted(self, "estimator_weights_")
X = self._validate_X_predict(X)
for i, _ in enumerate(self.estimators_, 1):
yield self._get_median_predict(X, limit=i)
| bsd-3-clause |
almarklein/scikit-image | doc/examples/plot_regionprops.py | 2 | 1300 | """
=========================
Measure region properties
=========================
This example shows how to measure properties of labelled image regions.
"""
import math
import matplotlib.pyplot as plt
import numpy as np
from skimage.draw import ellipse
from skimage.morphology import label
from skimage.measure import regionprops
from skimage.transform import rotate
image = np.zeros((600, 600))
rr, cc = ellipse(300, 350, 100, 220)
image[rr,cc] = 1
image = rotate(image, angle=15, order=0)
label_img = label(image)
regions = regionprops(label_img)
plt.imshow(image)
for props in regions:
y0, x0 = props.centroid
orientation = props.orientation
x1 = x0 + math.cos(orientation) * 0.5 * props.major_axis_length
y1 = y0 - math.sin(orientation) * 0.5 * props.major_axis_length
x2 = x0 - math.sin(orientation) * 0.5 * props.minor_axis_length
y2 = y0 - math.cos(orientation) * 0.5 * props.minor_axis_length
plt.plot((x0, x1), (y0, y1), '-r', linewidth=2.5)
plt.plot((x0, x2), (y0, y2), '-r', linewidth=2.5)
plt.plot(x0, y0, '.g', markersize=15)
minr, minc, maxr, maxc = props.bbox
bx = (minc, maxc, maxc, minc, minc)
by = (minr, minr, maxr, maxr, minr)
plt.plot(bx, by, '-b', linewidth=2.5)
plt.gray()
plt.axis((0, 600, 600, 0))
plt.show()
| bsd-3-clause |
ZenDevelopmentSystems/scikit-learn | benchmarks/bench_lasso.py | 297 | 3305 | """
Benchmarks of Lasso vs LassoLars
First, we fix a training set and increase the number of
samples. Then we plot the computation time as function of
the number of samples.
In the second benchmark, we increase the number of dimensions of the
training set. Then we plot the computation time as function of
the number of dimensions.
In both cases, only 10% of the features are informative.
"""
import gc
from time import time
import numpy as np
from sklearn.datasets.samples_generator import make_regression
def compute_bench(alpha, n_samples, n_features, precompute):
lasso_results = []
lars_lasso_results = []
it = 0
for ns in n_samples:
for nf in n_features:
it += 1
print('==================')
print('Iteration %s of %s' % (it, max(len(n_samples),
len(n_features))))
print('==================')
n_informative = nf // 10
X, Y, coef_ = make_regression(n_samples=ns, n_features=nf,
n_informative=n_informative,
noise=0.1, coef=True)
X /= np.sqrt(np.sum(X ** 2, axis=0)) # Normalize data
gc.collect()
print("- benchmarking Lasso")
clf = Lasso(alpha=alpha, fit_intercept=False,
precompute=precompute)
tstart = time()
clf.fit(X, Y)
lasso_results.append(time() - tstart)
gc.collect()
print("- benchmarking LassoLars")
clf = LassoLars(alpha=alpha, fit_intercept=False,
normalize=False, precompute=precompute)
tstart = time()
clf.fit(X, Y)
lars_lasso_results.append(time() - tstart)
return lasso_results, lars_lasso_results
if __name__ == '__main__':
from sklearn.linear_model import Lasso, LassoLars
import pylab as pl
alpha = 0.01 # regularization parameter
n_features = 10
list_n_samples = np.linspace(100, 1000000, 5).astype(np.int)
lasso_results, lars_lasso_results = compute_bench(alpha, list_n_samples,
[n_features], precompute=True)
pl.figure('scikit-learn LASSO benchmark results')
pl.subplot(211)
pl.plot(list_n_samples, lasso_results, 'b-',
label='Lasso')
pl.plot(list_n_samples, lars_lasso_results, 'r-',
label='LassoLars')
pl.title('precomputed Gram matrix, %d features, alpha=%s' % (n_features, alpha))
pl.legend(loc='upper left')
pl.xlabel('number of samples')
pl.ylabel('Time (s)')
pl.axis('tight')
n_samples = 2000
list_n_features = np.linspace(500, 3000, 5).astype(np.int)
lasso_results, lars_lasso_results = compute_bench(alpha, [n_samples],
list_n_features, precompute=False)
pl.subplot(212)
pl.plot(list_n_features, lasso_results, 'b-', label='Lasso')
pl.plot(list_n_features, lars_lasso_results, 'r-', label='LassoLars')
pl.title('%d samples, alpha=%s' % (n_samples, alpha))
pl.legend(loc='upper left')
pl.xlabel('number of features')
pl.ylabel('Time (s)')
pl.axis('tight')
pl.show()
| bsd-3-clause |
ifding/ifding.github.io | stylegan2-ada/metrics/clustering.py | 1 | 4125 | # Copyright (c) 2020, NVIDIA CORPORATION. All rights reserved.
#
# NVIDIA CORPORATION and its licensors retain all intellectual property
# and proprietary rights in and to this software, related documentation
# and any modifications thereto. Any use, reproduction, disclosure or
# distribution of this software and related documentation without an express
# license agreement from NVIDIA CORPORATION is strictly prohibited.
"""Inception Score (IS) from the paper
"Improved techniques for training GANs"."""
import pickle
import numpy as np
import tensorflow as tf
import dnnlib
import dnnlib.tflib as tflib
from metrics import metric_base
from training import dataset
from sklearn.metrics.cluster import normalized_mutual_info_score, adjusted_rand_score
def compute_purity(y_pred, y_true):
"""
Calculate the purity, a measurement of quality for the clustering
results.
Each cluster is assigned to the class which is most frequent in the
cluster. Using these classes, the percent accuracy is then calculated.
Returns:
A number between 0 and 1. Poor clusterings have a purity close to 0
while a perfect clustering has a purity of 1.
"""
# get the set of unique cluster ids
clusters = set(y_pred)
# find out what class is most frequent in each cluster
cluster_classes = {}
correct = 0
for cluster in clusters:
# get the indices of rows in this cluster
indices = np.where(y_pred == cluster)[0]
cluster_labels = y_true[indices]
majority_label = np.argmax(np.bincount(cluster_labels))
correct += np.sum(cluster_labels == majority_label)
#cor = np.sum(cluster_labels == majority_label)
#print(cluster, len(indices), float(cor)/len(indices))
return float(correct) / len(y_pred)
#----------------------------------------------------------------------------
class CL(metric_base.MetricBase):
def __init__(self, num_images, num_splits, minibatch_per_gpu, **kwargs):
super().__init__(**kwargs)
self.num_images = num_images
self.num_splits = num_splits
self.minibatch_per_gpu = minibatch_per_gpu
def _evaluate(self, E, G_kwargs, num_gpus, **_kwargs): # pylint: disable=arguments-differ
minibatch_size = num_gpus * self.minibatch_per_gpu
dataset_obj = dataset.load_dataset(**self._dataset_args)
dataset_obj.configure(minibatch_size)
trues = np.empty([self.num_images, 10], dtype=np.int32)
preds = np.empty([self.num_images, 10], dtype=np.float32)
# Construct TensorFlow graph.
result_expr = []
true_labels = []
for gpu_idx in range(num_gpus):
with tf.device(f'/gpu:{gpu_idx}'):
E_clone = E.clone()
images, labels = dataset_obj.get_minibatch_tf()
outputs = E_clone.get_output_for(images, labels, **G_kwargs)
output_logits = outputs[:, 512:]
output_labels = tf.nn.softmax(output_logits)
result_expr.append(output_labels)
true_labels.append(labels)
# Calculate activations for fakes.
for begin in range(0, self.num_images, minibatch_size):
self._report_progress(begin, self.num_images)
end = min(begin + minibatch_size, self.num_images)
trues[begin:end] = np.concatenate(tflib.run(true_labels), axis=0)[:end-begin]
preds[begin:end] = np.concatenate(tflib.run(result_expr), axis=0)[:end-begin]
labels_true = np.argmax(trues, axis=1)
labels_pred = np.argmax(preds, axis=1)
purity = compute_purity(labels_pred, labels_true)
ari = adjusted_rand_score(labels_true, labels_pred)
nmi = normalized_mutual_info_score(labels_true, labels_pred)
self._report_result(purity, suffix='purity')
self._report_result(ari, suffix='ari')
self._report_result(nmi, suffix='nmi')
#----------------------------------------------------------------------------
| mit |
huzq/scikit-learn | examples/applications/plot_outlier_detection_wine.py | 17 | 5819 | """
====================================
Outlier detection on a real data set
====================================
This example illustrates the need for robust covariance estimation
on a real data set. It is useful both for outlier detection and for
a better understanding of the data structure.
We selected two sets of two variables from the Boston housing data set
as an illustration of what kind of analysis can be done with several
outlier detection tools. For the purpose of visualization, we are working
with two-dimensional examples, but one should be aware that things are
not so trivial in high-dimension, as it will be pointed out.
In both examples below, the main result is that the empirical covariance
estimate, as a non-robust one, is highly influenced by the heterogeneous
structure of the observations. Although the robust covariance estimate is
able to focus on the main mode of the data distribution, it sticks to the
assumption that the data should be Gaussian distributed, yielding some biased
estimation of the data structure, but yet accurate to some extent.
The One-Class SVM does not assume any parametric form of the data distribution
and can therefore model the complex shape of the data much better.
First example
-------------
The first example illustrates how the Minimum Covariance Determinant
robust estimator can help concentrate on a relevant cluster when outlying
points exist. Here the empirical covariance estimation is skewed by points
outside of the main cluster. Of course, some screening tools would have pointed
out the presence of two clusters (Support Vector Machines, Gaussian Mixture
Models, univariate outlier detection, ...). But had it been a high-dimensional
example, none of these could be applied that easily.
"""
print(__doc__)
# Author: Virgile Fritsch <virgile.fritsch@inria.fr>
# License: BSD 3 clause
import numpy as np
from sklearn.covariance import EllipticEnvelope
from sklearn.svm import OneClassSVM
import matplotlib.pyplot as plt
import matplotlib.font_manager
from sklearn.datasets import load_wine
# Define "classifiers" to be used
classifiers = {
"Empirical Covariance": EllipticEnvelope(support_fraction=1.,
contamination=0.25),
"Robust Covariance (Minimum Covariance Determinant)":
EllipticEnvelope(contamination=0.25),
"OCSVM": OneClassSVM(nu=0.25, gamma=0.35)}
colors = ['m', 'g', 'b']
legend1 = {}
legend2 = {}
# Get data
X1 = load_wine()['data'][:, [1, 2]] # two clusters
# Learn a frontier for outlier detection with several classifiers
xx1, yy1 = np.meshgrid(np.linspace(0, 6, 500), np.linspace(1, 4.5, 500))
for i, (clf_name, clf) in enumerate(classifiers.items()):
plt.figure(1)
clf.fit(X1)
Z1 = clf.decision_function(np.c_[xx1.ravel(), yy1.ravel()])
Z1 = Z1.reshape(xx1.shape)
legend1[clf_name] = plt.contour(
xx1, yy1, Z1, levels=[0], linewidths=2, colors=colors[i])
legend1_values_list = list(legend1.values())
legend1_keys_list = list(legend1.keys())
# Plot the results (= shape of the data points cloud)
plt.figure(1) # two clusters
plt.title("Outlier detection on a real data set (wine recognition)")
plt.scatter(X1[:, 0], X1[:, 1], color='black')
bbox_args = dict(boxstyle="round", fc="0.8")
arrow_args = dict(arrowstyle="->")
plt.annotate("outlying points", xy=(4, 2),
xycoords="data", textcoords="data",
xytext=(3, 1.25), bbox=bbox_args, arrowprops=arrow_args)
plt.xlim((xx1.min(), xx1.max()))
plt.ylim((yy1.min(), yy1.max()))
plt.legend((legend1_values_list[0].collections[0],
legend1_values_list[1].collections[0],
legend1_values_list[2].collections[0]),
(legend1_keys_list[0], legend1_keys_list[1], legend1_keys_list[2]),
loc="upper center",
prop=matplotlib.font_manager.FontProperties(size=11))
plt.ylabel("ash")
plt.xlabel("malic_acid")
plt.show()
# %%
# Second example
# --------------
# The second example shows the ability of the Minimum Covariance Determinant
# robust estimator of covariance to concentrate on the main mode of the data
# distribution: the location seems to be well estimated, although the
# covariance is hard to estimate due to the banana-shaped distribution. Anyway,
# we can get rid of some outlying observations. The One-Class SVM is able to
# capture the real data structure, but the difficulty is to adjust its kernel
# bandwidth parameter so as to obtain a good compromise between the shape of
# the data scatter matrix and the risk of over-fitting the data.
# Get data
X2 = load_wine()['data'][:, [6, 9]] # "banana"-shaped
# Learn a frontier for outlier detection with several classifiers
xx2, yy2 = np.meshgrid(np.linspace(-1, 5.5, 500), np.linspace(-2.5, 19, 500))
for i, (clf_name, clf) in enumerate(classifiers.items()):
plt.figure(2)
clf.fit(X2)
Z2 = clf.decision_function(np.c_[xx2.ravel(), yy2.ravel()])
Z2 = Z2.reshape(xx2.shape)
legend2[clf_name] = plt.contour(
xx2, yy2, Z2, levels=[0], linewidths=2, colors=colors[i])
legend2_values_list = list(legend2.values())
legend2_keys_list = list(legend2.keys())
# Plot the results (= shape of the data points cloud)
plt.figure(2) # "banana" shape
plt.title("Outlier detection on a real data set (wine recognition)")
plt.scatter(X2[:, 0], X2[:, 1], color='black')
plt.xlim((xx2.min(), xx2.max()))
plt.ylim((yy2.min(), yy2.max()))
plt.legend((legend2_values_list[0].collections[0],
legend2_values_list[1].collections[0],
legend2_values_list[2].collections[0]),
(legend2_keys_list[0], legend2_keys_list[1], legend2_keys_list[2]),
loc="upper center",
prop=matplotlib.font_manager.FontProperties(size=11))
plt.ylabel("color_intensity")
plt.xlabel("flavanoids")
plt.show()
| bsd-3-clause |
lamastex/scalable-data-science | dbcArchives/2021/000_6-sds-3-x-dl/055_DLbyABr_04-ConvolutionalNetworks.py | 1 | 22551 | # Databricks notebook source
# MAGIC %md
# MAGIC ScaDaMaLe Course [site](https://lamastex.github.io/scalable-data-science/sds/3/x/) and [book](https://lamastex.github.io/ScaDaMaLe/index.html)
# MAGIC
# MAGIC This is a 2019-2021 augmentation and update of [Adam Breindel](https://www.linkedin.com/in/adbreind)'s initial notebooks.
# MAGIC
# MAGIC _Thanks to [Christian von Koch](https://www.linkedin.com/in/christianvonkoch/) and [William Anzén](https://www.linkedin.com/in/william-anz%C3%A9n-b52003199/) for their contributions towards making these materials Spark 3.0.1 and Python 3+ compliant._
# COMMAND ----------
# MAGIC %md
# MAGIC # Convolutional Neural Networks
# MAGIC ## aka CNN, ConvNet
# COMMAND ----------
# MAGIC %md
# MAGIC As a baseline, let's start a lab running with what we already know.
# MAGIC
# MAGIC We'll take our deep feed-forward multilayer perceptron network, with ReLU activations and reasonable initializations, and apply it to learning the MNIST digits.
# MAGIC
# MAGIC The main part of the code looks like the following (full code you can run is in the next cell):
# MAGIC
# MAGIC ```
# MAGIC # imports, setup, load data sets
# MAGIC
# MAGIC model = Sequential()
# MAGIC model.add(Dense(20, input_dim=784, kernel_initializer='normal', activation='relu'))
# MAGIC model.add(Dense(15, kernel_initializer='normal', activation='relu'))
# MAGIC model.add(Dense(10, kernel_initializer='normal', activation='softmax'))
# MAGIC model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['categorical_accuracy'])
# MAGIC
# MAGIC categorical_labels = to_categorical(y_train, num_classes=10)
# MAGIC
# MAGIC history = model.fit(X_train, categorical_labels, epochs=100, batch_size=100)
# MAGIC
# MAGIC # print metrics, plot errors
# MAGIC ```
# MAGIC
# MAGIC Note the changes, which are largely about building a classifier instead of a regression model:
# MAGIC * Output layer has one neuron per category, with softmax activation
# MAGIC * __Loss function is cross-entropy loss__
# MAGIC * Accuracy metric is categorical accuracy
# COMMAND ----------
# MAGIC %md
# MAGIC Let's hold pointers into wikipedia for these new concepts.
# COMMAND ----------
# MAGIC %scala
# MAGIC //This allows easy embedding of publicly available information into any other notebook
# MAGIC //Example usage:
# MAGIC // displayHTML(frameIt("https://en.wikipedia.org/wiki/Latent_Dirichlet_allocation#Topics_in_LDA",250))
# MAGIC def frameIt( u:String, h:Int ) : String = {
# MAGIC """<iframe
# MAGIC src=""""+ u+""""
# MAGIC width="95%" height="""" + h + """"
# MAGIC sandbox>
# MAGIC <p>
# MAGIC <a href="http://spark.apache.org/docs/latest/index.html">
# MAGIC Fallback link for browsers that, unlikely, don't support frames
# MAGIC </a>
# MAGIC </p>
# MAGIC </iframe>"""
# MAGIC }
# MAGIC displayHTML(frameIt("https://en.wikipedia.org/wiki/Cross_entropy#Cross-entropy_error_function_and_logistic_regression",500))
# COMMAND ----------
# MAGIC %scala
# MAGIC displayHTML(frameIt("https://en.wikipedia.org/wiki/Softmax_function",380))
# COMMAND ----------
# MAGIC %md
# MAGIC The following is from: [https://www.quora.com/How-does-Keras-calculate-accuracy](https://www.quora.com/How-does-Keras-calculate-accuracy).
# MAGIC
# MAGIC **Categorical accuracy:**
# MAGIC
# MAGIC ```%python
# MAGIC def categorical_accuracy(y_true, y_pred):
# MAGIC return K.cast(K.equal(K.argmax(y_true, axis=-1),
# MAGIC K.argmax(y_pred, axis=-1)),
# MAGIC K.floatx())
# MAGIC ```
# MAGIC
# MAGIC > `K.argmax(y_true)` takes the highest value to be the prediction and matches against the comparative set.
# COMMAND ----------
# MAGIC %md
# MAGIC Watch (1:39)
# MAGIC * [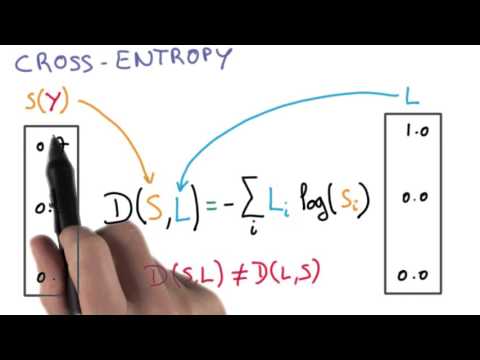](https://www.youtube.com/watch?v=tRsSi_sqXjI)
# MAGIC
# MAGIC Watch (1:54)
# MAGIC * [](https://www.youtube.com/watch?v=x449QQDhMDE)
# COMMAND ----------
from keras.models import Sequential
from keras.layers import Dense
from keras.utils import to_categorical
import sklearn.datasets
import datetime
import matplotlib.pyplot as plt
import numpy as np
train_libsvm = "/dbfs/databricks-datasets/mnist-digits/data-001/mnist-digits-train.txt"
test_libsvm = "/dbfs/databricks-datasets/mnist-digits/data-001/mnist-digits-test.txt"
X_train, y_train = sklearn.datasets.load_svmlight_file(train_libsvm, n_features=784)
X_train = X_train.toarray()
X_test, y_test = sklearn.datasets.load_svmlight_file(test_libsvm, n_features=784)
X_test = X_test.toarray()
model = Sequential()
model.add(Dense(20, input_dim=784, kernel_initializer='normal', activation='relu'))
model.add(Dense(15, kernel_initializer='normal', activation='relu'))
model.add(Dense(10, kernel_initializer='normal', activation='softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['categorical_accuracy'])
categorical_labels = to_categorical(y_train, num_classes=10)
start = datetime.datetime.today()
history = model.fit(X_train, categorical_labels, epochs=40, batch_size=100, validation_split=0.1, verbose=2)
scores = model.evaluate(X_test, to_categorical(y_test, num_classes=10))
print
for i in range(len(model.metrics_names)):
print("%s: %f" % (model.metrics_names[i], scores[i]))
print ("Start: " + str(start))
end = datetime.datetime.today()
print ("End: " + str(end))
print ("Elapse: " + str(end-start))
# COMMAND ----------
# MAGIC %md
# MAGIC after about a minute we have:
# MAGIC
# MAGIC ```
# MAGIC ...
# MAGIC
# MAGIC Epoch 40/40
# MAGIC 1s - loss: 0.0610 - categorical_accuracy: 0.9809 - val_loss: 0.1918 - val_categorical_accuracy: 0.9583
# MAGIC
# MAGIC ...
# MAGIC
# MAGIC loss: 0.216120
# MAGIC
# MAGIC categorical_accuracy: 0.955000
# MAGIC
# MAGIC Start: 2017-12-06 07:35:33.948102
# MAGIC
# MAGIC End: 2017-12-06 07:36:27.046130
# MAGIC
# MAGIC Elapse: 0:00:53.098028
# MAGIC ```
# COMMAND ----------
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
fig.set_size_inches((5,5))
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'val'], loc='upper left')
display(fig)
# COMMAND ----------
# MAGIC %md
# MAGIC What are the big takeaways from this experiment?
# MAGIC
# MAGIC 1. We get pretty impressive "apparent error" accuracy right from the start! A small network gets us to training accuracy 97% by epoch 20
# MAGIC 2. The model *appears* to continue to learn if we let it run, although it does slow down and oscillate a bit.
# MAGIC 3. Our test accuracy is about 95% after 5 epochs and never gets better ... it gets worse!
# MAGIC 4. Therefore, we are overfitting very quickly... most of the "training" turns out to be a waste.
# MAGIC 5. For what it's worth, we get 95% accuracy without much work.
# MAGIC
# MAGIC This is not terrible compared to other, non-neural-network approaches to the problem. After all, we could probably tweak this a bit and do even better.
# MAGIC
# MAGIC But we talked about using deep learning to solve "95%" problems or "98%" problems ... where one error in 20, or 50 simply won't work. If we can get to "multiple nines" of accuracy, then we can do things like automate mail sorting and translation, create cars that react properly (all the time) to street signs, and control systems for robots or drones that function autonomously.
# MAGIC
# MAGIC Try two more experiments (try them separately):
# MAGIC 1. Add a third, hidden layer.
# MAGIC 2. Increase the size of the hidden layers.
# MAGIC
# MAGIC Adding another layer slows things down a little (why?) but doesn't seem to make a difference in accuracy.
# MAGIC
# MAGIC Adding a lot more neurons into the first topology slows things down significantly -- 10x as many neurons, and only a marginal increase in accuracy. Notice also (in the plot) that the learning clearly degrades after epoch 50 or so.
# MAGIC
# MAGIC ... We need a new approach!
# MAGIC
# MAGIC ---
# MAGIC
# MAGIC ... let's think about this:
# MAGIC
# MAGIC ### What is layer 2 learning from layer 1? Combinations of pixels
# MAGIC
# MAGIC #### Combinations of pixels contain information but...
# MAGIC
# MAGIC There are a lot of them (combinations) and they are "fragile"
# MAGIC
# MAGIC In fact, in our last experiment, we basically built a model that memorizes a bunch of "magic" pixel combinations.
# MAGIC
# MAGIC What might be a better way to build features?
# MAGIC
# MAGIC * When humans perform this task, we look not at arbitrary pixel combinations, but certain geometric patterns -- lines, curves, loops.
# MAGIC * These features are made up of combinations of pixels, but they are far from arbitrary
# MAGIC * We identify these features regardless of translation, rotation, etc.
# MAGIC
# MAGIC Is there a way to get the network to do the same thing?
# MAGIC
# MAGIC I.e., in layer one, identify pixels. Then in layer 2+, identify abstractions over pixels that are translation-invariant 2-D shapes?
# MAGIC
# MAGIC We could look at where a "filter" that represents one of these features (e.g., and edge) matches the image.
# MAGIC
# MAGIC How would this work?
# MAGIC
# MAGIC ### Convolution
# MAGIC
# MAGIC Convolution in the general mathematical sense is define as follows:
# MAGIC
# MAGIC <img src="https://i.imgur.com/lurC2Cx.png" width=300>
# MAGIC
# MAGIC The convolution we deal with in deep learning is a simplified case. We want to compare two signals. Here are two visualizations, courtesy of Wikipedia, that help communicate how convolution emphasizes features:
# MAGIC
# MAGIC <img src="http://i.imgur.com/EDCaMl2.png" width=500>
# MAGIC
# MAGIC ---
# MAGIC
# MAGIC #### Here's an animation (where we change \\({\tau}\\))
# MAGIC <img src="http://i.imgur.com/0BFcnaw.gif">
# MAGIC
# MAGIC __In one sense, the convolution captures and quantifies the pattern matching over space__
# MAGIC
# MAGIC If we perform this in two dimensions, we can achieve effects like highlighting edges:
# MAGIC
# MAGIC <img src="http://i.imgur.com/DKEXIII.png">
# MAGIC
# MAGIC The matrix here, also called a convolution kernel, is one of the functions we are convolving. Other convolution kernels can blur, "sharpen," etc.
# MAGIC
# MAGIC ### So we'll drop in a number of convolution kernels, and the network will learn where to use them? Nope. Better than that.
# MAGIC
# MAGIC ## We'll program in the *idea* of discrete convolution, and the network will learn what kernels extract meaningful features!
# MAGIC
# MAGIC The values in a (fixed-size) convolution kernel matrix will be variables in our deep learning model. Although inuitively it seems like it would be hard to learn useful params, in fact, since those variables are used repeatedly across the image data, it "focuses" the error on a smallish number of parameters with a lot of influence -- so it should be vastly *less* expensive to train than just a huge fully connected layer like we discussed above.
# MAGIC
# MAGIC This idea was developed in the late 1980s, and by 1989, Yann LeCun (at AT&T/Bell Labs) had built a practical high-accuracy system (used in the 1990s for processing handwritten checks and mail).
# MAGIC
# MAGIC __How do we hook this into our neural networks?__
# MAGIC
# MAGIC * First, we can preserve the geometric properties of our data by "shaping" the vectors as 2D instead of 1D.
# MAGIC
# MAGIC * Then we'll create a layer whose value is not just activation applied to weighted sum of inputs, but instead it's the result of a dot-product (element-wise multiply and sum) between the kernel and a patch of the input vector (image).
# MAGIC * This value will be our "pre-activation" and optionally feed into an activation function (or "detector")
# MAGIC
# MAGIC <img src="http://i.imgur.com/ECyi9lL.png">
# MAGIC
# MAGIC
# MAGIC If we perform this operation at lots of positions over the image, we'll get lots of outputs, as many as one for every input pixel.
# MAGIC
# MAGIC
# MAGIC <img src="http://i.imgur.com/WhOrJ0Y.jpg">
# MAGIC
# MAGIC * So we'll add another layer that "picks" the highest convolution pattern match from nearby pixels, which
# MAGIC * makes our pattern match a little bit translation invariant (a fuzzy location match)
# MAGIC * reduces the number of outputs significantly
# MAGIC * This layer is commonly called a pooling layer, and if we pick the "maximum match" then it's a "max pooling" layer.
# MAGIC
# MAGIC <img src="http://i.imgur.com/9iPpfpb.png">
# MAGIC
# MAGIC __The end result is that the kernel or filter together with max pooling creates a value in a subsequent layer which represents the appearance of a pattern in a local area in a prior layer.__
# MAGIC
# MAGIC __Again, the network will be given a number of "slots" for these filters and will learn (by minimizing error) what filter values produce meaningful features. This is the key insight into how modern image-recognition networks are able to generalize -- i.e., learn to tell 6s from 7s or cats from dogs.__
# MAGIC
# MAGIC <img src="http://i.imgur.com/F8eH3vj.png">
# MAGIC
# MAGIC ## Ok, let's build our first ConvNet:
# MAGIC
# MAGIC First, we want to explicity shape our data into a 2-D configuration. We'll end up with a 4-D tensor where the first dimension is the training examples, then each example is 28x28 pixels, and we'll explicitly say it's 1-layer deep. (Why? with color images, we typically process over 3 or 4 channels in this last dimension)
# MAGIC
# MAGIC A step by step animation follows:
# MAGIC * http://cs231n.github.io/assets/conv-demo/index.html
# COMMAND ----------
train_libsvm = "/dbfs/databricks-datasets/mnist-digits/data-001/mnist-digits-train.txt"
test_libsvm = "/dbfs/databricks-datasets/mnist-digits/data-001/mnist-digits-test.txt"
X_train, y_train = sklearn.datasets.load_svmlight_file(train_libsvm, n_features=784)
X_train = X_train.toarray()
X_test, y_test = sklearn.datasets.load_svmlight_file(test_libsvm, n_features=784)
X_test = X_test.toarray()
X_train = X_train.reshape( (X_train.shape[0], 28, 28, 1) )
X_train = X_train.astype('float32')
X_train /= 255
y_train = to_categorical(y_train, num_classes=10)
X_test = X_test.reshape( (X_test.shape[0], 28, 28, 1) )
X_test = X_test.astype('float32')
X_test /= 255
y_test = to_categorical(y_test, num_classes=10)
# COMMAND ----------
# MAGIC %md
# MAGIC Now the model:
# COMMAND ----------
from keras.layers import Dense, Dropout, Activation, Flatten, Conv2D, MaxPooling2D
model = Sequential()
model.add(Conv2D(8, # number of kernels
(4, 4), # kernel size
padding='valid', # no padding; output will be smaller than input
input_shape=(28, 28, 1)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2,2)))
model.add(Flatten())
model.add(Dense(128))
model.add(Activation('relu')) # alternative syntax for applying activation
model.add(Dense(10))
model.add(Activation('softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
# COMMAND ----------
# MAGIC %md
# MAGIC ... and the training loop and output:
# COMMAND ----------
start = datetime.datetime.today()
history = model.fit(X_train, y_train, batch_size=128, epochs=8, verbose=2, validation_split=0.1)
scores = model.evaluate(X_test, y_test, verbose=1)
print
for i in range(len(model.metrics_names)):
print("%s: %f" % (model.metrics_names[i], scores[i]))
# COMMAND ----------
fig, ax = plt.subplots()
fig.set_size_inches((5,5))
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'val'], loc='upper left')
display(fig)
# COMMAND ----------
# MAGIC %md
# MAGIC ### Our MNIST ConvNet
# MAGIC
# MAGIC In our first convolutional MNIST experiment, we get to almost 99% validation accuracy in just a few epochs (a minutes or so on CPU)!
# MAGIC
# MAGIC The training accuracy is effectively 100%, though, so we've almost completely overfit (i.e., memorized the training data) by this point and need to do a little work if we want to keep learning.
# MAGIC
# MAGIC Let's add another convolutional layer:
# COMMAND ----------
model = Sequential()
model.add(Conv2D(8, # number of kernels
(4, 4), # kernel size
padding='valid',
input_shape=(28, 28, 1)))
model.add(Activation('relu'))
model.add(Conv2D(8, (4, 4)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2,2)))
model.add(Flatten())
model.add(Dense(128))
model.add(Activation('relu'))
model.add(Dense(10))
model.add(Activation('softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
history = model.fit(X_train, y_train, batch_size=128, epochs=15, verbose=2, validation_split=0.1)
scores = model.evaluate(X_test, y_test, verbose=1)
print
for i in range(len(model.metrics_names)):
print("%s: %f" % (model.metrics_names[i], scores[i]))
# COMMAND ----------
# MAGIC %md
# MAGIC While that's running, let's look at a number of "famous" convolutional networks!
# MAGIC
# MAGIC ### LeNet (Yann LeCun, 1998)
# MAGIC
# MAGIC <img src="http://i.imgur.com/k5hMtMK.png">
# MAGIC
# MAGIC <img src="http://i.imgur.com/ERV9pHW.gif">
# COMMAND ----------
# MAGIC %md <img src="http://i.imgur.com/TCN9C4P.png">
# COMMAND ----------
# MAGIC %md
# MAGIC ### AlexNet (2012)
# MAGIC
# MAGIC <img src="http://i.imgur.com/CpokDKV.jpg">
# MAGIC
# MAGIC <img src="http://i.imgur.com/Ld2QhXr.jpg">
# COMMAND ----------
# MAGIC %md
# MAGIC ### Back to our labs: Still Overfitting
# MAGIC
# MAGIC We're making progress on our test error -- about 99% -- but just a bit for all the additional time, due to the network overfitting the data.
# MAGIC
# MAGIC There are a variety of techniques we can take to counter this -- forms of regularization.
# MAGIC
# MAGIC Let's try a relatively simple solution solution that works surprisingly well: add a pair of `Dropout` filters, a layer that randomly omits a fraction of neurons from each training batch (thus exposing each neuron to only part of the training data).
# MAGIC
# MAGIC We'll add more convolution kernels but shrink them to 3x3 as well.
# COMMAND ----------
model = Sequential()
model.add(Conv2D(32, # number of kernels
(3, 3), # kernel size
padding='valid',
input_shape=(28, 28, 1)))
model.add(Activation('relu'))
model.add(Conv2D(32, (3, 3)))
model.add(Activation('relu'))
model.add(MaxPooling2D(pool_size=(2,2)))
model.add(Dropout(rate=1-0.25)) # <- regularize, new parameter rate added (rate=1-keep_prob)
model.add(Flatten())
model.add(Dense(128))
model.add(Activation('relu'))
model.add(Dropout(rate=1-0.5)) # <-regularize, new parameter rate added (rate=1-keep_prob)
model.add(Dense(10))
model.add(Activation('softmax'))
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
history = model.fit(X_train, y_train, batch_size=128, epochs=15, verbose=2)
scores = model.evaluate(X_test, y_test, verbose=2)
print
for i in range(len(model.metrics_names)):
print("%s: %f" % (model.metrics_names[i], scores[i]))
# COMMAND ----------
# MAGIC %md
# MAGIC While that's running, let's look at some more recent ConvNet architectures:
# MAGIC
# MAGIC ### VGG16 (2014)
# MAGIC
# MAGIC <img src="http://i.imgur.com/gl4kZDf.png">
# COMMAND ----------
# MAGIC %md
# MAGIC ### GoogLeNet (2014)
# MAGIC
# MAGIC <img src="http://i.imgur.com/hvmtDqN.png">
# MAGIC
# MAGIC *"Inception" layer: parallel convolutions at different resolutions*
# MAGIC
# MAGIC ### Residual Networks (2015-)
# MAGIC
# MAGIC Skip layers to improve training (error propagation). Residual layers learn from details at multiple previous layers.
# MAGIC
# MAGIC <img src="http://i.imgur.com/32g8Ykl.png">
# COMMAND ----------
# MAGIC %md
# MAGIC ---
# MAGIC
# MAGIC > __ASIDE: Atrous / Dilated Convolutions__
# MAGIC
# MAGIC > An atrous or dilated convolution is a convolution filter with "holes" in it. Effectively, it is a way to enlarge the filter spatially while not adding as many parameters or attending to every element in the input.
# MAGIC
# MAGIC > Why? Covering a larger input volume allows recognizing coarser-grained patterns; restricting the number of parameters is a way of regularizing or constraining the capacity of the model, making training easier.
# MAGIC
# MAGIC ---
# COMMAND ----------
# MAGIC %md
# MAGIC ## *Lab Wrapup*
# MAGIC
# MAGIC From the last lab, you should have a test accuracy of over 99.1%
# MAGIC
# MAGIC For one more activity, try changing the optimizer to old-school "sgd" -- just to see how far we've come with these modern gradient descent techniques in the last few years.
# MAGIC
# MAGIC Accuracy will end up noticeably worse ... about 96-97% test accuracy. Two key takeaways:
# MAGIC
# MAGIC * Without a good optimizer, even a very powerful network design may not achieve results
# MAGIC * In fact, we could replace the word "optimizer" there with
# MAGIC * initialization
# MAGIC * activation
# MAGIC * regularization
# MAGIC * (etc.)
# MAGIC * All of these elements we've been working with operate together in a complex way to determine final performance
# COMMAND ----------
# MAGIC %md
# MAGIC Of course this world evolves fast - see the new kid in the CNN block -- **capsule networks**
# MAGIC
# MAGIC > Hinton: “The pooling operation used in convolutional neural networks is a big mistake and the fact that it works so well is a disaster.”
# MAGIC
# MAGIC Well worth the 8 minute read:
# MAGIC * [https://medium.com/ai%C2%B3-theory-practice-business/understanding-hintons-capsule-networks-part-i-intuition-b4b559d1159b](https://medium.com/ai%C2%B3-theory-practice-business/understanding-hintons-capsule-networks-part-i-intuition-b4b559d1159b)
# MAGIC
# MAGIC To understand deeper:
# MAGIC * original paper: [https://arxiv.org/abs/1710.09829](https://arxiv.org/abs/1710.09829)
# MAGIC
# MAGIC [Keras capsule network example](https://keras.io/examples/cifar10_cnn_capsule/)
# COMMAND ----------
# MAGIC %md
# MAGIC # More resources
# MAGIC
# MAGIC - http://www.wildml.com/2015/12/implementing-a-cnn-for-text-classification-in-tensorflow/
# MAGIC - https://openai.com/
# COMMAND ----------
| unlicense |
3DGenomes/tadbit | _pytadbit/mapping/analyze.py | 1 | 67679 | """
18 Nov 2014
"""
from warnings import warn
from collections import OrderedDict
from pysam import AlignmentFile
from scipy.stats import norm as sc_norm, skew, kurtosis
from scipy.stats import pearsonr, spearmanr, linregress
from scipy.sparse.linalg import eigsh
from numpy.linalg import eigh
import numpy as np
try:
from matplotlib import rcParams
from matplotlib import pyplot as plt
from matplotlib.backends.backend_pdf import PdfPages
from matplotlib.colors import LinearSegmentedColormap
except ImportError:
warn('matplotlib not found\n')
from pytadbit import HiC_data
from pytadbit.utils.extraviews import tadbit_savefig, setup_plot
from pytadbit.utils.tadmaths import nozero_log_matrix as nozero_log
from pytadbit.utils.tadmaths import right_double_mad as mad
from pytadbit.parsers.hic_parser import load_hic_data_from_reads
from pytadbit.utils.extraviews import nicer
from pytadbit.utils.file_handling import mkdir
def hic_map(data, resolution=None, normalized=False, masked=None,
by_chrom=False, savefig=None, show=False, savedata=None,
focus=None, clim=None, perc_clim=None, cmap='jet', pdf=False, decay=True,
perc=20, name=None, decay_resolution=None, **kwargs):
"""
function to retrieve data from HiC-data object. Data can be stored as
a square matrix, or drawn using matplotlib
:param data: can be either a path to a file with pre-processed reads
(filtered or not), or a Hi-C-data object
:param None resolution: at which to bin the data (try having a dense matrix
with < 10% of cells with zero interaction counts). Note: not necessary
if a hic_data object is passed as 'data'.
:param False normalized: used normalized data, based on precalculated biases
:param masked: a list of columns to be removed. Usually because to few
interactions
:param False by_chrom: data can be stored in a partitioned way. This
parameter can take the values of:
* 'intra': one output per each chromosome will be created
* 'inter': one output per each possible pair of chromosome will be
created
* 'all' : both of the above outputs
:param None savefig: path where to store the output images. Note that, if
the by_chrom option is used, then savefig will be the name of the
directory containing the output files.
:param None savedata: path where to store the output matrices. Note that, if
the by_chrom option is used, then savefig will be the name of the
directory containing the output files.
:param None focus: can be either two number (i.e.: (1, 100)) specifying the
start and end position of the sub-matrix to display (start and end, along
the diagonal of the original matrix); or directly a chromosome name; or
two chromosome names (i.e.: focus=('chr2, chrX')), in order to store the
data corresponding to inter chromosomal interactions between these two
chromosomes
:param True decay: plot the correlation between genomic distance and
interactions (usually a decay).
:param False force_image: force to generate an image even if resolution is
crazy...
:param None clim: cutoff for the upper and lower bound in the coloring scale
of the heatmap. (perc_clim should be set to None)
:param None perc_clim: cutoff for the upper and lower bound in the coloring scale
of the heatmap; in percentile. (clim should be set to None)
:param False pdf: when using the bny_chrom option, to specify the format of
the stored images
:param jet cmap: color map to be used for the heatmap; "tadbit" color map is
also implemented and will use percentiles of the distribution of
interactions to defines intensities of red.
:param None decay_resolution: chromatin fragment size to consider when
calculating decay of the number of interactions with genomic distance.
Default is equal to resolution of the matrix.
"""
if isinstance(data, str):
data = load_hic_data_from_reads(data, resolution=resolution, **kwargs)
if not kwargs.get('get_sections', True) and decay:
warn('WARNING: not decay not available when get_sections is off.')
decay = False
if clim and perc_clim:
raise Exception('ERROR: only one of clim or perc_clim should be set\n')
hic_data = data
resolution = data.resolution
if not decay_resolution:
decay_resolution = resolution
if hic_data.bads and not masked:
masked = hic_data.bads
# save and draw the data
if by_chrom:
if focus:
raise Exception('Incompatible options focus and by_chrom\n')
if savedata:
mkdir(savedata)
if savefig:
mkdir(savefig)
for i, crm1 in enumerate(hic_data.chromosomes):
for crm2 in hic_data.chromosomes.keys()[i:]:
if by_chrom == 'intra' and crm1 != crm2:
continue
if by_chrom == 'inter' and crm1 == crm2:
continue
try:
subdata = hic_data.get_matrix(focus=(crm1, crm2), normalized=normalized)
start1, _ = hic_data.section_pos[crm1]
start2, _ = hic_data.section_pos[crm2]
masked1 = {}
masked2 = {}
if focus and hic_data.bads:
# rescale masked
masked1 = dict([(m - start1, hic_data.bads[m])
for m in hic_data.bads])
masked2 = dict([(m - start2, hic_data.bads[m])
for m in hic_data.bads])
if masked1 or masked2:
for i in xrange(len(subdata)):
if i in masked1:
subdata[i] = [float('nan')
for j in xrange(len(subdata))]
for j in xrange(len(subdata)):
if j in masked2:
subdata[i][j] = float('nan')
if savedata:
hic_data.write_matrix('%s/%s.mat' % (
savedata, '_'.join(set((crm1, crm2)))),
focus=(crm1, crm2),
normalized=normalized)
if show or savefig:
if (len(subdata) > 10000
and not kwargs.get('force_image', False)):
warn('WARNING: Matrix image not created, more than '
'10000 rows, use a lower resolution to create images')
continue
draw_map(subdata,
OrderedDict([(k, hic_data.chromosomes[k])
for k in hic_data.chromosomes.keys()
if k in [crm1, crm2]]),
hic_data.section_pos,
'%s/%s.%s' % (savefig,
'_'.join(set((crm1, crm2))),
'pdf' if pdf else 'png'),
show, one=True, clim=clim, perc_clim=perc_clim,
cmap=cmap, decay_resolution=decay_resolution,
perc=perc, name=name, cistrans=float('NaN'))
except ValueError, e:
print 'Value ERROR: problem with chromosome %s' % crm1
print str(e)
except IndexError, e:
print 'Index ERROR: problem with chromosome %s' % crm1
print str(e)
else:
if savedata:
hic_data.write_matrix(savedata, focus=focus,
normalized=normalized)
if show or savefig:
subdata = hic_data.get_matrix(focus=focus, normalized=normalized)
if (len(subdata) > 10000 and not kwargs.get('force_image', False)):
warn('WARNING: Matrix image not created, more than '
'10000 rows, use a lower resolution to create images')
return
start1 = hic_data._focus_coords(focus)[0]
if focus and masked:
# rescale masked
masked = dict([(m - start1, masked[m]) for m in masked])
if masked:
for i in xrange(len(subdata)):
if i in masked:
subdata[i] = [float('nan')
for j in xrange(len(subdata))]
for j in xrange(len(subdata)):
if j in masked:
subdata[i][j] = float('nan')
draw_map(subdata,
{} if focus else hic_data.chromosomes,
hic_data.section_pos, savefig, show,
one = True if focus else False, decay=decay,
clim=clim, perc_clim=perc_clim, cmap=cmap,
decay_resolution=decay_resolution,
perc=perc, normalized=normalized,
max_diff=kwargs.get('max_diff', None),
name=name, cistrans=float('NaN') if focus else
hic_data.cis_trans_ratio(normalized,
kwargs.get('exclude', None),
kwargs.get('diagonal', True),
kwargs.get('equals', None)))
def draw_map(data, genome_seq, cumcs, savefig, show, one=False, clim=None,
perc_clim=None, cmap='jet', decay=False, perc=20, name=None,
cistrans=None, decay_resolution=10000, normalized=False,
max_diff=None):
_ = plt.figure(figsize=(15.,12.5))
if not max_diff:
max_diff = len(data)
ax1 = plt.axes([0.34, 0.08, 0.6, 0.7205])
ax2 = plt.axes([0.07, 0.65, 0.21, 0.15])
if decay:
ax3 = plt.axes([0.07, 0.42, 0.21, 0.15])
plot_distance_vs_interactions(data, genome_seq=genome_seq, axe=ax3,
resolution=decay_resolution,
max_diff=max_diff, normalized=normalized)
ax4 = plt.axes([0.34, 0.805, 0.6, 0.04], sharex=ax1)
ax5 = plt.axes([0.34, 0.845, 0.6, 0.04], sharex=ax1)
ax6 = plt.axes([0.34, 0.885, 0.6, 0.04], sharex=ax1)
try:
minoridata = np.nanmin(data)
maxoridata = np.nanmax(data)
except AttributeError:
vals = [i for d in data for i in d if not np.isnan(i)]
minoridata = np.min(vals)
maxoridata = np.max(vals)
totaloridata = np.nansum([data[i][j] for i in xrange(len(data))
for j in xrange(i, len(data[i]))]) # may not be square
data = nozero_log(data, np.log2)
vals = np.array([i for d in data for i in d])
vals = vals[np.isfinite(vals)]
if perc_clim:
try:
clim = np.percentile(vals, perc_clim[0]), np.percentile(vals, perc_clim[1])
except ValueError:
clim = None
mindata = np.nanmin(vals)
maxdata = np.nanmax(vals)
diff = maxdata - mindata
norm = lambda x: (x - mindata) / diff
posI = 0.01 if not clim else norm(clim[0]) if clim[0] != None else 0.01
posF = 1.0 if not clim else norm(clim[1]) if clim[1] != None else 1.0
if cmap == 'tadbit':
cuts = perc
cdict = {'red' : [(0.0, 1.0, 1.0)],
'green': [(0.0, 1.0, 1.0)],
'blue' : [(0.0, 1.0, 1.0)]}
for i in np.linspace(posI, posF, cuts, endpoint=False):
prc = (i / (posF - posI)) / 1.75
pos = norm(np.percentile(vals, i * 100.))
# print '%7.4f %7.4f %7.4f %7.4f' % (prc, pos, np.percentile(vals, i * 100.), i)
cdict['red' ].append([pos, 1 , 1 ])
cdict['green'].append([pos, 1 - prc, 1 - prc])
cdict['blue' ].append([pos, 1 - prc, 1 - prc])
cdict['red' ].append([1.0, 1, 1])
cdict['green'].append([1.0, 0, 0])
cdict['blue' ].append([1.0, 0, 0])
cmap = LinearSegmentedColormap(cmap, cdict)
clim = None
else:
cmap = plt.get_cmap(cmap)
cmap.set_bad('darkgrey', 1)
ax1.imshow(data, interpolation='none',
cmap=cmap, vmin=clim[0] if clim else None, vmax=clim[1] if clim else None)
size1 = len(data)
size2 = len(data[0])
if size1 == size2:
for i in xrange(size1):
for j in xrange(i, size2):
if np.isnan(data[i][j]):
data[i][j] = 0
data[j][i] = 0
else:
for i in xrange(size1):
for j in xrange(size2):
if np.isnan(data[i][j]):
data[i][j] = 0
#data[j][i] = data[i][j]
try:
evals, evect = eigh(data)
sort_perm = evals.argsort()
evect = evect[sort_perm]
except:
evals, evect = None, None
data = [i for d in data for i in d if np.isfinite(i)]
gradient = np.linspace(np.nanmin(data),
np.nanmax(data), max(size1, size2))
gradient = np.vstack((gradient, gradient))
try:
h = ax2.hist(data, color='darkgrey', linewidth=2,
bins=20, histtype='step', density=True)
except AttributeError:
h = ax2.hist(data, color='darkgrey', linewidth=2,
bins=20, histtype='step', normed=True)
_ = ax2.imshow(gradient, aspect='auto', cmap=cmap,
vmin=clim[0] if clim else None, vmax=clim[1] if clim else None,
extent=(np.nanmin(data), np.nanmax(data) , 0, max(h[0])))
if genome_seq:
for crm in genome_seq:
ax1.vlines([cumcs[crm][0]-.5, cumcs[crm][1]-.5], cumcs[crm][0]-.5, cumcs[crm][1]-.5,
color='w', linestyle='-', linewidth=1, alpha=1)
ax1.hlines([cumcs[crm][1]-.5, cumcs[crm][0]-.5], cumcs[crm][0]-.5, cumcs[crm][1]-.5,
color='w', linestyle='-', linewidth=1, alpha=1)
ax1.vlines([cumcs[crm][0]-.5, cumcs[crm][1]-.5], cumcs[crm][0]-.5, cumcs[crm][1]-.5,
color='k', linestyle='--')
ax1.hlines([cumcs[crm][1]-.5, cumcs[crm][0]-.5], cumcs[crm][0]-.5, cumcs[crm][1]-.5,
color='k', linestyle='--')
if not one:
vals = [0]
keys = ['']
for crm in genome_seq:
vals.append(cumcs[crm][0])
keys.append(crm)
vals.append(cumcs[crm][1])
ax1.set_yticks(vals)
ax1.set_yticklabels('')
ax1.set_yticks([float(vals[i]+vals[i+1])/2
for i in xrange(len(vals) - 1)], minor=True)
ax1.set_yticklabels(keys, minor=True)
for t in ax1.yaxis.get_minor_ticks():
t.tick1On = False
t.tick2On = False
# totaloridata = ''.join([j + ('' if (i+1)%3 else ',') for i, j in enumerate(str(totaloridata)[::-1])])[::-1].strip(',')
# minoridata = ''.join([j + ('' if (i+1)%3 else ',') for i, j in enumerate(str(minoridata)[::-1])])[::-1].strip(',')
# maxoridata = ''.join([j + ('' if (i+1)%3 else ',') for i, j in enumerate(str(maxoridata)[::-1])])[::-1].strip(',')
plt.figtext(0.05,0.25, ''.join([
(name + '\n') if name else '',
'Number of interactions: %s\n' % str(totaloridata),
('' if np.isnan(cistrans) else
('Percentage of cis interactions: %.0f%%\n' % (cistrans*100))),
'Min interactions: %s\n' % (minoridata),
'Max interactions: %s\n' % (maxoridata)]))
ax2.set_xlim((np.nanmin(data), np.nanmax(data)))
ax2.set_ylim((0, max(h[0])))
ax1.set_xlim ((-0.5, size1 - .5))
ax1.set_ylim ((-0.5, size2 - .5))
ax2.set_xlabel('log interaction count')
# we reduce the number of dots displayed.... we just want to see the shape
subdata = np.array(list(set([float(int(d*100))/100 for d in data])))
try:
normfit = sc_norm.pdf(subdata, np.nanmean(data), np.nanstd(data))
except AttributeError:
normfit = sc_norm.pdf(subdata, np.mean(data), np.std(data))
ax2.plot(subdata, normfit, 'w.', markersize=2.5, alpha=.4)
ax2.plot(subdata, normfit, 'k.', markersize=1.5, alpha=1)
ax2.set_title('skew: %.3f, kurtosis: %.3f' % (skew(data),
kurtosis(data)))
try:
ax4.vlines(range(size1), 0, evect[:,-1], color='k')
except (TypeError, IndexError):
pass
ax4.hlines(0, 0, size2, color='red')
ax4.set_ylabel('E1')
ax4.set_yticklabels([])
try:
ax5.vlines(range(size1), 0, evect[:,-2], color='k')
except (TypeError, IndexError):
pass
ax5.hlines(0, 0, size2, color='red')
ax5.set_ylabel('E2')
ax5.set_yticklabels([])
try:
ax6.vlines(range(size1), 0, evect[:,-3], color='k')
except (TypeError, IndexError):
pass
ax6.hlines(0, 0, size2, color='red')
ax6.set_ylabel('E3')
ax6.set_yticklabels([])
xticklabels = ax4.get_xticklabels() + ax5.get_xticklabels() + ax6.get_xticklabels()
plt.setp(xticklabels, visible=False)
if savefig:
tadbit_savefig(savefig)
elif show:
plt.show()
plt.close('all')
def plot_distance_vs_interactions(data, min_diff=1, max_diff=1000, show=False,
genome_seq=None, resolution=None, axe=None,
savefig=None, normalized=False,
plot_each_cell=False):
"""
Plot the number of interactions observed versus the genomic distance between
the mapped ends of the read. The slope is expected to be around -1, in
logarithmic scale and between 700 kb and 10 Mb (according to the prediction
of the fractal globule model).
:param data: input file name (either tsv or TADbit generated BAM), or
HiC_data object or list of lists
:param 10 min_diff: lower limit (in number of bins)
:param 1000 max_diff: upper limit (in number of bins) to look for
:param 100 resolution: group reads that are closer than this resolution
parameter
:param_hash False plot_each_cell: if false, only the mean distances by bin
will be represented, otherwise each pair of interactions will be plotted.
:param None axe: a matplotlib.axes.Axes object to define the plot
appearance
:param None savefig: path to a file where to save the image generated;
if None, the image will be shown using matplotlib GUI (the extension
of the file name will determine the desired format).
:returns: slope, intercept and R square of each of the 3 correlations
"""
if isinstance(data, basestring):
resolution = resolution or 1
dist_intr = dict([(i, {})
for i in xrange(min_diff, max_diff)])
fhandler = open(data)
line = fhandler.next()
while line.startswith('#'):
line = fhandler.next()
try:
while True:
_, cr1, ps1, _, _, _, _, cr2, ps2, _ = line.split('\t', 9)
if cr1 != cr2:
line = fhandler.next()
continue
diff = abs(int(ps1) / resolution - int(ps2) / resolution)
if max_diff > diff >= min_diff:
try:
dist_intr[diff][int(ps1) / resolution] += 1.
except KeyError:
dist_intr[diff][int(ps1) / resolution] = 1.
line = fhandler.next()
except StopIteration:
pass
fhandler.close()
for diff in dist_intr:
dist_intr[diff] = [dist_intr[diff].get(k, 0)
for k in xrange(max(dist_intr[diff]) - diff)]
elif isinstance(data, HiC_data):
resolution = resolution or data.resolution
dist_intr = dict([(i, []) for i in xrange(min_diff, max_diff)])
if normalized:
get_data = lambda x, y: data[x, y] / data.bias[x] / data.bias[y]
else:
get_data = lambda x, y: data[x, y]
max_diff = min(len(data), max_diff)
if data.section_pos:
for crm in data.section_pos:
for diff in xrange(min_diff, min(
(max_diff, 1 + data.chromosomes[crm]))):
for i in xrange(data.section_pos[crm][0],
data.section_pos[crm][1] - diff):
dist_intr[diff].append(get_data(i, i + diff))
else:
for diff in xrange(min_diff, max_diff):
for i in xrange(len(data) - diff):
if not np.isnan(data[i, i + diff]):
dist_intr[diff].append(get_data(i, diff))
elif isinstance(data, dict): # if we pass decay/expected dictionary, computes weighted mean
dist_intr = {}
for i in range(min_diff, max_diff):
val = [data[c][i] for c in data
if i in data[c] and data[c][i] != data[c].get(i-1, 0)]
if val:
dist_intr[i] = [sum(val) / float(len(val))]
else:
dist_intr[i] = [0]
else:
dist_intr = dict([(i, []) for i in xrange(min_diff, max_diff)])
if genome_seq:
max_diff = min(max(genome_seq.values()), max_diff)
cnt = 0
for crm in genome_seq:
for diff in xrange(min_diff, min(
(max_diff, genome_seq[crm]))):
for i in xrange(cnt, cnt + genome_seq[crm] - diff):
if not np.isnan(data[i][i + diff]):
dist_intr[diff].append(data[i][i + diff])
cnt += genome_seq[crm]
else:
max_diff = min(len(data), max_diff)
for diff in xrange(min_diff, max_diff):
for i in xrange(len(data) - diff):
if not np.isnan(data[i][i + diff]):
dist_intr[diff].append(data[i][i + diff])
resolution = resolution or 1
if not axe:
fig=plt.figure()
axe = fig.add_subplot(111)
# remove last part of the plot in case no interaction is count... reduce max_dist
for diff in xrange(max_diff - 1, min_diff, -1):
try:
if not dist_intr[diff]:
del(dist_intr[diff])
max_diff -=1
continue
except KeyError:
max_diff -=1
continue
break
# get_cmap the mean values perc bins
mean_intr = dict([(i, float(sum(dist_intr[i])) / len(dist_intr[i]))
for i in dist_intr if len(dist_intr[i])])
if plot_each_cell:
xp, yp = [], []
for x, y in sorted(dist_intr.items(), key=lambda x:x[0]):
xp.extend([x] * len(y))
yp.extend(y)
x = []
y = []
for k in xrange(len(xp)):
if yp[k]:
x.append(xp[k])
y.append(yp[k])
axe.plot(x, y, color='grey', marker='.', alpha=0.1, ms=1,
linestyle='None')
xp, yp = zip(*sorted(mean_intr.items(), key=lambda x:x[0]))
x = []
y = []
for k in xrange(len(xp)):
if yp[k]:
x.append(xp[k])
y.append(yp[k])
axe.plot(x, y, 'k.', alpha=0.4)
best = (float('-inf'), 0, 0, 0, 0, 0, 0, 0, 0, 0)
logx = np.log(x)
logy = np.log(y)
ntries = 100
# set k for better fit
# for k in xrange(1, ntries/5, ntries/5/5):
if resolution == 1:
k = 1
for i in xrange(3, ntries-2-k):
v1 = i * len(x) / ntries
try:
a1, b1, r21, _, _ = linregress(logx[ :v1], logy[ :v1])
except ValueError:
a1 = b1 = r21 = 0
r21 *= r21
for j in xrange(i + 1 + k, ntries - 2 - k):
v2 = j * len(x) / ntries
try:
a2, b2, r22, _, _ = linregress(logx[v1+k:v2], logy[v1+k:v2])
a3, b3, r23, _, _ = linregress(logx[v2+k: ], logy[v2+k: ])
except ValueError:
a2 = b2 = r22 = 0
a3 = b3 = r23 = 0
r2 = r21 + r22**2 + r23**2
if r2 > best[0]:
best = (r2, v1, v2, a1, a2, a3,
b1, b2, b3, k)
# plot line of best fit
(v1, v2,
a1, a2, a3,
b1, b2, b3, k) = best[1:]
yfit1 = lambda xx: np.exp(b1 + a1*np.array (np.log(xx)))
yfit2 = lambda xx: np.exp(b2 + a2*np.array (np.log(xx)))
yfit3 = lambda xx: np.exp(b3 + a3*np.array (np.log(xx)))
axe.plot(x[ :v1], yfit1(x[ :v1] ), color= 'yellow', lw=2,
label = r'$\alpha_{%s}=%.2f$' % (
'0-0.7 \mathrm{ Mb}' if resolution != 1 else '1', a1))
#label = r'$\alpha_1=%.2f$ (0-%d)' % (a1, x[v1]))
axe.plot(x[v1+k:v2], yfit2(x[v1+k:v2]), color= 'orange', lw=2,
label = r'$\alpha_{%s}=%.2f$' % (
'0.7-10 \mathrm{ Mb}' if resolution != 1 else '2', a2))
# label = r'$\alpha_2=%.2f$ (%d-%d)' % (a2, x[v1], x[v2]))
axe.plot(x[v2+k: ], yfit3(x[v2+k: ] ), color= 'red' , lw=2,
label = r'$\alpha_{%s}=%.2f$' % (
'10 \mathrm{ Mb}-\infty' if resolution != 1 else '3', a3))
# label = r'$\alpha_3=%.2f$ (%d-$\infty$)' % (a3, x[v2+k]))
else:
# from 0.7 Mb
v1 = 700000 / resolution
# to 10 Mb
v2 = 10000000 / resolution
try:
a1, b1, r21, _, _ = linregress(logx[ :v1], logy[ :v1])
except ValueError:
a1, b1, r21 = 0, 0, 0
try:
a2, b2, r22, _, _ = linregress(logx[v1:v2], logy[v1:v2])
except ValueError:
a2, b2, r22 = 0, 0, 0
try:
a3, b3, r23, _, _ = linregress(logx[v2: ], logy[v2: ])
except ValueError:
a3, b3, r23 = 0, 0, 0
yfit1 = lambda xx: np.exp(b1 + a1*np.array (np.log(xx)))
yfit2 = lambda xx: np.exp(b2 + a2*np.array (np.log(xx)))
yfit3 = lambda xx: np.exp(b3 + a3*np.array (np.log(xx)))
axe.plot(x[ :v1], yfit1(x[ :v1] ), color= 'yellow', lw=2,
label = r'$\alpha_{%s}=%.2f$' % (
'0-0.7 \mathrm{ Mb}' if resolution != 1 else '1', a1))
#label = r'$\alpha_1=%.2f$ (0-%d)' % (a1, x[v1]))
axe.plot(x[v1:v2], yfit2(x[v1:v2]), color= 'orange', lw=2,
label = r'$\alpha_{%s}=%.2f$' % (
'0.7-10 \mathrm{ Mb}' if resolution != 1 else '2', a2))
# label = r'$\alpha_2=%.2f$ (%d-%d)' % (a2, x[v1], x[v2]))
axe.plot(x[v2: ], yfit3(x[v2: ] ), color= 'red' , lw=2,
label = r'$\alpha_{%s}=%.2f$' % (
'10 \mathrm{ Mb}-\infty' if resolution != 1 else '3', a3))
# label = r'$\alpha_3=%.2f$ (%d-$\infty$)' % (a3, x[v2+k]))
axe.set_ylabel('Log interaction count')
axe.set_xlabel('Log genomic distance (resolution: %s)' % nicer(resolution))
axe.legend(loc='lower left', frameon=False)
axe.set_xscale('log')
axe.set_yscale('log')
axe.set_xlim((min_diff, max_diff))
try:
axe.set_ylim((0, max(y)))
except ValueError:
pass
if savefig:
tadbit_savefig(savefig)
plt.close('all')
elif show:
plt.show()
plt.close('all')
return (a1, b1, r21), (a2, b2, r22), (a3, b3, r23)
def plot_iterative_mapping(fnam1, fnam2, total_reads=None, axe=None, savefig=None):
"""
Plots the number of reads mapped at each step of the mapping process (in the
case of the iterative mapping, each step is mapping process with a given
size of fragments).
:param fnam: input file name
:param total_reads: total number of reads in the initial FASTQ file
:param None axe: a matplotlib.axes.Axes object to define the plot
appearance
:param None savefig: path to a file where to save the image generated;
if None, the image will be shown using matplotlib GUI (the extension
of the file name will determine the desired format).
:returns: a dictionary with the number of reads per mapped length
"""
count_by_len = {}
total_reads = total_reads or 1
if not axe:
fig=plt.figure()
_ = fig.add_subplot(111)
colors = ['olive', 'darkcyan']
iteration = False
for i, fnam in enumerate([fnam1, fnam2]):
fhandler = open(fnam)
line = fhandler.next()
count_by_len[i] = {}
while line.startswith('#'):
if line.startswith('# MAPPED '):
itr, num = line.split()[2:]
count_by_len[i][int(itr)] = int(num)
line = fhandler.next()
if not count_by_len[i]:
iteration = True
try:
while True:
_, length, _, _ = line.rsplit('\t', 3)
try:
count_by_len[i][int(length)] += 1
except KeyError:
count_by_len[i][int(length)] = 1
line = fhandler.next()
except StopIteration:
pass
fhandler.close()
lengths = sorted(count_by_len[i].keys())
for k in lengths[::-1]:
count_by_len[i][k] += sum([count_by_len[i][j]
for j in lengths if j < k])
plt.plot(lengths, [float(count_by_len[i][l]) / total_reads
for l in lengths],
label='read' + str(i + 1), linewidth=2, color=colors[i])
if iteration:
plt.xlabel('read length (bp)')
else:
plt.xlabel('Iteration number')
if total_reads != 1:
plt.ylabel('Proportion of mapped reads')
else:
plt.ylabel('Number of mapped reads')
plt.legend(loc=4)
if savefig:
tadbit_savefig(savefig)
elif not axe:
plt.show()
plt.close('all')
return count_by_len
def fragment_size(fnam, savefig=None, nreads=None, max_size=99.9, axe=None,
show=False, xlog=False, stats=('median', 'perc_max'),
too_large=10000):
"""
Plots the distribution of dangling-ends lengths
:param fnam: input file name
:param None savefig: path where to store the output images.
:param 99.9 max_size: top percentage of distances to consider, within the
top 0.01% are usually found very long outliers.
:param False xlog: represent x axis in logarithmic scale
:param ('median', 'perc_max') stats: returns this set of values calculated from the
distribution of insert/fragment sizes. Possible values are:
- 'median' median of the distribution
- 'mean' mean of the distribution
- 'perc_max' percentil defined by the other parameter 'max_size'
- 'first_deacay' starting from the median of the distribution to the
first window where 10 consecutive insert sizes are counted less than
a given value (this given value is equal to the sum of all
sizes divided by 100 000)
- 'MAD' Double Median Adjusted Deviation
:param 10000 too_large: upper bound limit for fragment size to consider
:param None nreads: number of reads to process (default: all reads)
:returns: the median value and the percentile inputed as max_size.
"""
distr = {}
genome_seq = OrderedDict()
pos = 0
fhandler = open(fnam)
for line in fhandler:
if line.startswith('#'):
if line.startswith('# CRM '):
crm, clen = line[6:].split('\t')
genome_seq[crm] = int(clen)
else:
break
pos += len(line)
fhandler.seek(pos)
des = []
for line in fhandler:
(crm1, pos1, dir1, _, re1, _,
crm2, pos2, dir2, _, re2) = line.strip().split('\t')[1:12]
if re1 == re2 and crm1 == crm2 and dir1 == '1' and dir2 == '0':
pos1, pos2 = int(pos1), int(pos2)
des.append(pos2 - pos1)
if len(des) == nreads:
break
des = [i for i in des if i <= too_large]
fhandler.close()
if not des:
raise Exception('ERROR: no dangling-ends found in %s' % (fnam))
max_perc = np.percentile(des, max_size)
perc99 = np.percentile(des, 99)
perc01 = np.percentile(des, 1)
perc50 = np.percentile(des, 50)
meanfr = np.mean(des)
perc95 = np.percentile(des, 95)
perc05 = np.percentile(des, 5)
to_return = {'median': perc50}
cutoff = len(des) / 100000.
count = 0
for v in xrange(int(perc50), int(max(des))):
if des.count(v) < cutoff:
count += 1
else:
count = 0
if count >= 10:
to_return['first_decay'] = v - 10
break
else:
raise Exception('ERROR: not found')
to_return['perc_max'] = max_perc
to_return['MAD'] = mad(des)
to_return['mean'] = meanfr
if not savefig and not axe and not show:
return [to_return[k] for k in stats]
ax = setup_plot(axe, figsize=(10, 5.5))
desapan = ax.axvspan(perc95, perc99, facecolor='black', alpha=.2,
label='1-99%% DEs\n(%.0f-%.0f nts)' % (perc01, perc99))
ax.axvspan(perc01, perc05, facecolor='black', alpha=.2)
desapan = ax.axvspan(perc05, perc95, facecolor='black', alpha=.4,
label='5-95%% DEs\n(%.0f-%.0f nts)' % (perc05, perc95))
deshist = ax.hist(des, bins=100, range=(0, max_perc), lw=2,
alpha=.5, edgecolor='darkred', facecolor='darkred', label='Dangling-ends')
ylims = ax.get_ylim()
plots = []
ax.set_xlabel('Genomic distance between reads')
ax.set_ylabel('Count')
ax.set_title('Distribution of dangling-ends ' +
'lenghts\nmedian: %s (mean: %s), top %.1f%%: %0.f nts' % (
int(perc50), int(meanfr), max_size, int(max_perc)))
if xlog:
ax.set_xscale('log')
ax.set_xlim((50, max_perc))
plt.subplots_adjust(left=0.1, right=0.75)
ax.legend(bbox_to_anchor=(1.4, 1), frameon=False)
if savefig:
tadbit_savefig(savefig)
elif show and not axe:
plt.show()
plt.close('all')
return [to_return[k] for k in stats]
def plot_genomic_distribution(fnam, first_read=None, resolution=10000,
ylim=None, yscale=None, savefig=None, show=False,
savedata=None, chr_names=None, nreads=None):
"""
Plot the number of reads in bins along the genome (or along a given
chromosome).
:param fnam: input file name
:param True first_read: uses first read.
:param 100 resolution: group reads that are closer than this resolution
parameter
:param None ylim: a tuple of lower and upper bound for the y axis
:param None yscale: if set_bad to "log" values will be represented in log2
scale
:param None savefig: path to a file where to save the image generated;
if None, the image will be shown using matplotlib GUI (the extension
of the file name will determine the desired format).
:param None savedata: path where to store the output read counts per bin.
:param None chr_names: can pass a list of chromosome names in case only some
them the need to be plotted (this option may last even more than default)
:param None nreads: number of reads to process (default: all reads)
"""
if first_read:
warn('WARNING: first_read parameter should no loonger be used.')
distr = {}
genome_seq = OrderedDict()
if chr_names:
chr_names = set(chr_names)
cond1 = lambda x: x not in chr_names
else:
cond1 = lambda x: False
if nreads:
cond2 = lambda x: x >= nreads
else:
cond2 = lambda x: False
cond = lambda x, y: cond1(x) or cond2(y)
count = 0
pos = 0
fhandler = open(fnam)
for line in fhandler:
if line.startswith('#'):
if line.startswith('# CRM '):
crm, clen = line[6:].split('\t')
genome_seq[crm] = int(clen)
else:
break
pos += len(line)
fhandler.seek(pos)
for line in fhandler:
line = line.strip().split('\t')
count += 1
for idx1, idx2 in ((1, 3), (7, 9)):
crm, pos = line[idx1:idx2]
if cond(crm, count):
if cond2(count):
break
continue
pos = int(pos) / resolution
try:
distr[crm][pos] += 1
except KeyError:
try:
distr[crm][pos] = 1
except KeyError:
distr[crm] = {pos: 1}
else:
continue
break
fhandler.close()
if savefig or show:
_ = plt.figure(figsize=(15, 1 + 3 * len(
chr_names if chr_names else distr.keys())))
max_y = max([max(distr[c].values()) for c in distr])
max_x = max([len(distr[c].values()) for c in distr])
ncrms = len(chr_names if chr_names else genome_seq if genome_seq else distr)
data = {}
for i, crm in enumerate(chr_names if chr_names else genome_seq
if genome_seq else distr):
try:
# data[crm] = [distr[crm].get(j, 0) for j in xrange(max(distr[crm]))] # genome_seq[crm]
data[crm] = [distr[crm].get(j, 0)
for j in xrange(genome_seq[crm] / resolution + 1)]
if savefig or show:
plt.subplot(ncrms, 1, i + 1)
plt.plot(range(genome_seq[crm] / resolution + 1), data[crm],
color='red', lw=1.5, alpha=0.7)
if yscale:
plt.yscale(yscale)
except KeyError:
pass
if savefig or show:
if ylim:
plt.vlines(genome_seq[crm] / resolution, ylim[0], ylim[1])
else:
plt.vlines(genome_seq[crm] / resolution, 0, max_y)
plt.xlim((0, max_x))
plt.ylim(ylim or (0, max_y))
plt.title(crm)
if savefig:
tadbit_savefig(savefig)
if not show:
plt.close('all')
elif show:
plt.show()
if savedata:
out = open(savedata, 'w')
out.write('# CRM\tstart-end\tcount\n')
out.write('\n'.join('%s\t%d-%d\t%d' % (c, (i * resolution) + 1,
((i + 1) * resolution), v)
for c in data for i, v in enumerate(data[c])))
out.write('\n')
out.close()
def _unitize(vals):
return np.argsort(vals) / float(len(vals))
def correlate_matrices(hic_data1, hic_data2, max_dist=10, intra=False, axe=None,
savefig=None, show=False, savedata=None, min_dist=1,
normalized=False, remove_bad_columns=True, **kwargs):
"""
Compare the interactions of two Hi-C matrices at a given distance,
with Spearman rank correlation.
Also computes the SCC reproducibility score as in HiCrep (see
https://doi.org/10.1101/gr.220640.117). It's implementation is inspired
by the version implemented in dryhic by Enrique Vidal
(https://github.com/qenvio/dryhic).
:param hic_data1: Hi-C-data object
:param hic_data2: Hi-C-data object
:param 1 resolution: to be used for scaling the plot
:param 10 max_dist: maximum distance from diagonal (e.g. 10 mean we will
not look further than 10 times the resolution)
:param 1 min_dist: minimum distance from diagonal (set to 0 to reproduce
result from HicRep)
:param None savefig: path to save the plot
:param False intra: only takes into account intra-chromosomal contacts
:param False show: displays the plot
:param False normalized: use normalized data
:param True remove_bads: computes the union of bad columns between samples
and exclude them from the comparison
:returns: list of correlations, list of genomic distances, SCC and standard
deviation of SCC
"""
spearmans = []
pearsons = []
dists = []
weigs = []
if normalized:
get_the_guy1 = lambda i, j: (hic_data1[j, i] / hic_data1.bias[i] /
hic_data1.bias[j])
get_the_guy2 = lambda i, j: (hic_data2[j, i] / hic_data2.bias[i] /
hic_data2.bias[j])
else:
get_the_guy1 = lambda i, j: hic_data1[j, i]
get_the_guy2 = lambda i, j: hic_data2[j, i]
if remove_bad_columns:
# union of bad columns
bads = hic_data1.bads.copy()
bads.update(hic_data2.bads)
if (intra and hic_data1.sections and hic_data2.sections and
hic_data1.sections == hic_data2.sections):
for dist in xrange(1, max_dist + 1):
diag1 = []
diag2 = []
for crm in hic_data1.section_pos:
for j in xrange(hic_data1.section_pos[crm][0],
hic_data1.section_pos[crm][1] - dist):
i = j + dist
if j in bads or i in bads:
continue
diag1.append(get_the_guy1(i, j))
diag2.append(get_the_guy2(i, j))
spearmans.append(spearmanr(diag1, diag2)[0])
pearsons.append(spearmanr(diag1, diag2)[0])
r1 = _unitize(diag1)
r2 = _unitize(diag2)
weigs.append((np.var(r1, ddof=1) *
np.var(r2, ddof=1))**0.5 * len(diag1))
dists.append(dist)
else:
if intra:
warn('WARNING: hic_dta does not contain chromosome coordinates, ' +
'intra set to False')
for dist in xrange(min_dist, max_dist + min_dist):
diag1 = []
diag2 = []
for j in xrange(len(hic_data1) - dist):
i = j + dist
if j in bads or i in bads:
continue
diag1.append(get_the_guy1(i, j))
diag2.append(get_the_guy2(i, j))
spearmans.append(spearmanr(diag1, diag2)[0])
pearsons.append(pearsonr(diag1, diag2)[0])
r1 = _unitize(diag1)
r2 = _unitize(diag2)
weigs.append((np.var(r1, ddof=1) *
np.var(r2, ddof=1))**0.5 * len(diag1))
dists.append(dist)
# compute scc
# print pearsons
# print weigs
tot_weigth = sum(weigs)
scc = sum(pearsons[i] * weigs[i] / tot_weigth
for i in xrange(len(pearsons)))
var_corr = np.var(pearsons, ddof=1)
std = (sum(weigs[i]**2 for i in xrange(len(pearsons))) * var_corr /
sum(weigs)**2)**0.5
# plot
if show or savefig or axe:
if not axe:
fig = plt.figure()
axe = fig.add_subplot(111)
given_axe = False
else:
given_axe = True
axe.plot(dists, spearmans, color='orange', linewidth=3, alpha=.8)
axe.set_xlabel('Genomic distance in bins')
axe.set_ylabel('Spearman rank correlation')
axe.set_xlim((0, dists[-1]))
if savefig:
tadbit_savefig(savefig)
if show:
plt.show()
if not given_axe:
plt.close('all')
if savedata:
out = open(savedata, 'w')
out.write('# genomic distance\tSpearman rank correlation\n')
for i in xrange(len(spearmans)):
out.write('%s\t%s\n' % (dists[i], spearmans[i]))
out.close()
if kwargs.get('get_bads', False):
return spearmans, dists, scc, std, bads
return spearmans, dists, scc, std
def _evec_dist(v1,v2):
d1=np.dot(v1-v2,v1-v2)
d2=np.dot(v1+v2,v1+v2)
if d1<d2:
d=d1
else:
d=d2
return np.sqrt(d)
def _get_Laplacian(M):
S=M.sum(1)
i_nz=np.where(S>0)[0]
S=S[i_nz]
M=(M[i_nz].T)[i_nz].T
S=1/np.sqrt(S)
M=S*M
M=(S*M.T).T
n=np.size(S)
M=np.identity(n)-M
M=(M+M.T)/2
return M
def get_ipr(evec):
ipr=1.0/(evec*evec*evec*evec).sum()
return ipr
def get_reproducibility(hic_data1, hic_data2, num_evec, verbose=True,
normalized=False, remove_bad_columns=True):
"""
Compute reproducibility score similarly to HiC-spector
(https://doi.org/10.1093/bioinformatics/btx152)
:param hic_data1: Hi-C-data object
:param hic_data2: Hi-C-data object
:param 20 num_evec: number of eigenvectors to compare
:returns: reproducibility score (bellow 0.5 ~ different cell types)
"""
M1 = hic_data1.get_matrix(normalized=normalized)
M2 = hic_data2.get_matrix(normalized=normalized)
if remove_bad_columns:
# union of bad columns
bads = hic_data1.bads.copy()
bads.update(hic_data2.bads)
# remove them form both matrices
for bad in sorted(bads, reverse=True):
del(M1[bad])
del(M2[bad])
for i in xrange(len(M1)):
_ = M1[i].pop(bad)
_ = M2[i].pop(bad)
M1 = np.matrix(M1)
M2 = np.matrix(M2)
k1=np.sign(M1.A).sum(1)
d1=np.diag(M1.A)
kd1=~((k1==1)*(d1>0))
k2=np.sign(M2.A).sum(1)
d2=np.diag(M2.A)
kd2=~((k2==1)*(d2>0))
iz=np.nonzero((k1+k2>0)*(kd1>0)*(kd2>0))[0]
M1b=(M1[iz].A.T)[iz].T
M2b=(M2[iz].A.T)[iz].T
i_nz1=np.where(M1b.sum(1)>0)[0]
i_nz2=np.where(M2b.sum(1)>0)[0]
i_z1=np.where(M1b.sum(1)==0)[0]
i_z2=np.where(M2b.sum(1)==0)[0]
M1b_L=_get_Laplacian(M1b)
M2b_L=_get_Laplacian(M2b)
a1, b1=eigsh(M1b_L,k=num_evec,which="SM")
a2, b2=eigsh(M2b_L,k=num_evec,which="SM")
b1_extend=np.zeros((np.size(M1b,0),num_evec))
b2_extend=np.zeros((np.size(M2b,0),num_evec))
for i in range(num_evec):
b1_extend[i_nz1,i]=b1[:,i]
b2_extend[i_nz2,i]=b2[:,i]
ipr_cut=5
ipr1=np.zeros(num_evec)
ipr2=np.zeros(num_evec)
for i in range(num_evec):
ipr1[i]=get_ipr(b1_extend[:,i])
ipr2[i]=get_ipr(b2_extend[:,i])
b1_extend_eff=b1_extend[:,ipr1>ipr_cut]
b2_extend_eff=b2_extend[:,ipr2>ipr_cut]
num_evec_eff=min(np.size(b1_extend_eff,1),np.size(b2_extend_eff,1))
evd=np.zeros(num_evec_eff)
for i in range(num_evec_eff):
evd[i]=_evec_dist(b1_extend_eff[:,i],b2_extend_eff[:,i])
Sd=evd.sum()
l=np.sqrt(2)
evs=abs(l-Sd/num_evec_eff)/l
N = float(M1.shape[1])
if verbose:
if (np.sum(ipr1>N/100)<=1)|(np.sum(ipr2>N/100)<=1):
print("at least one of the maps does not look like typical Hi-C maps")
else:
print("size of maps: %d" %(np.size(M1,0)))
print("reproducibility score: %6.3f " %(evs))
print("num_evec_eff: %d" %(num_evec_eff))
return evs
def eig_correlate_matrices(hic_data1, hic_data2, nvect=6, normalized=False,
savefig=None, show=False, savedata=None,
remove_bad_columns=True, **kwargs):
"""
Compare the interactions of two Hi-C matrices using their 6 first
eigenvectors, with Pearson correlation
:param hic_data1: Hi-C-data object
:param hic_data2: Hi-C-data object
:param 6 nvect: number of eigenvectors to compare
:param None savefig: path to save the plot
:param False show: displays the plot
:param False normalized: use normalized data
:param True remove_bads: computes the union of bad columns between samples
and exclude them from the comparison
:param kwargs: any argument to pass to matplotlib imshow function
:returns: matrix of correlations
"""
data1 = hic_data1.get_matrix(normalized=normalized)
data2 = hic_data2.get_matrix(normalized=normalized)
## reduce matrices to remove bad columns
if remove_bad_columns:
# union of bad columns
bads = hic_data1.bads.copy()
bads.update(hic_data2.bads)
# remove them form both matrices
for bad in sorted(bads, reverse=True):
del(data1[bad])
del(data2[bad])
for i in xrange(len(data1)):
_ = data1[i].pop(bad)
_ = data2[i].pop(bad)
# get the log
data1 = nozero_log(data1, np.log2)
data2 = nozero_log(data2, np.log2)
# get the eigenvectors
ev1, evect1 = eigh(data1)
ev2, evect2 = eigh(data2)
corr = [[0 for _ in xrange(nvect)] for _ in xrange(nvect)]
# sort eigenvectors according to their eigenvalues => first is last!!
sort_perm = ev1.argsort()
ev1.sort()
evect1 = evect1[sort_perm]
sort_perm = ev2.argsort()
ev2.sort()
evect2 = evect2[sort_perm]
# calculate Pearson correlation
for i in xrange(nvect):
for j in xrange(nvect):
corr[i][j] = abs(pearsonr(evect1[:,-i-1],
evect2[:,-j-1])[0])
# plot
axe = plt.axes([0.1, 0.1, 0.6, 0.8])
cbaxes = plt.axes([0.85, 0.1, 0.03, 0.8])
if show or savefig:
im = axe.imshow(corr, interpolation="nearest",origin='lower', **kwargs)
axe.set_xlabel('Eigen Vectors exp. 1')
axe.set_ylabel('Eigen Vectors exp. 2')
axe.set_xticks(range(nvect))
axe.set_yticks(range(nvect))
axe.set_xticklabels(range(1, nvect + 2))
axe.set_yticklabels(range(1, nvect + 2))
axe.xaxis.set_tick_params(length=0, width=0)
axe.yaxis.set_tick_params(length=0, width=0)
cbar = plt.colorbar(im, cax = cbaxes )
cbar.ax.set_ylabel('Pearson correlation', rotation=90*3,
verticalalignment='bottom')
axe2 = axe.twinx()
axe2.set_yticks(range(nvect))
axe2.set_yticklabels(['%.1f' % (e) for e in ev2[-nvect:][::-1]])
axe2.set_ylabel('corresponding Eigen Values exp. 2', rotation=90*3,
verticalalignment='bottom')
axe2.set_ylim((-0.5, nvect - 0.5))
axe2.yaxis.set_tick_params(length=0, width=0)
axe3 = axe.twiny()
axe3.set_xticks(range(nvect))
axe3.set_xticklabels(['%.1f' % (e) for e in ev1[-nvect:][::-1]])
axe3.set_xlabel('corresponding Eigen Values exp. 1')
axe3.set_xlim((-0.5, nvect - 0.5))
axe3.xaxis.set_tick_params(length=0, width=0)
axe.set_ylim((-0.5, nvect - 0.5))
axe.set_xlim((-0.5, nvect - 0.5))
if savefig:
tadbit_savefig(savefig)
if show:
plt.show()
plt.close('all')
if savedata:
out = open(savedata, 'w')
out.write('# ' + '\t'.join(['Eigen Vector %s'% i
for i in xrange(nvect)]) + '\n')
for i in xrange(nvect):
out.write('\t'.join([str(corr[i][j])
for j in xrange(nvect)]) + '\n')
out.close()
if kwargs.get('get_bads', False):
return corr, bads
else:
return corr
def plot_rsite_reads_distribution(reads_file, outprefix, window=20,
maxdist=1000):
de_right={}
de_left={}
print "process reads"
fl=open(reads_file)
while True:
line=fl.next()
if not line.startswith('#'):
break
nreads=0
try:
while True:
nreads += 1
if nreads % 1000000 == 0:
print nreads
try:
_, n1, sb1, sd1, l1, ru1, rd1, n2, sb2, sd2, l2, ru2, rd2\
= line.split()
sb1, sd1, l1, ru1, rd1, sb2, sd2, l2, ru2, rd2 = \
map(int, [sb1, sd1, l1, ru1, rd1, sb2, sd2, l2,
ru2, rd2])
except ValueError:
print line
raise ValueError("line is not the right format!")
if n1 != n2:
line=fl.next()
continue
#read1 ahead of read2
if sb1 > sb2:
sb1, sd1, l1, ru1, rd1, sb2, sd2, l2, ru2, rd2 = \
sb2, sd2, l2, ru2, rd2, sb1, sd1, l1, ru1, rd1
#direction always -> <-
if not (sd1 == 1 and sd2 == 0):
line=fl.next()
continue
#close to the diagonal
if sb2-sb1 > maxdist:
line=fl.next()
continue
#close to RE 1
if abs(sb1-ru1) < abs(sb1-rd1):
rc1=ru1
else:
rc1=rd1
pos=sb1-rc1
if abs(pos)<=window:
if not pos in de_right:
de_right[pos]=0
de_right[pos]+=1
#close to RE 2
if abs(sb2-ru2) < abs(sb2-rd2):
rc2=ru2
else:
rc2=rd2
pos=sb2-rc2
if abs(pos)<=window:
if not pos in de_left:
de_left[pos]=0
de_left[pos]+=1
line=fl.next()
except StopIteration:
pass
print " finished processing {} reads".format(nreads)
#transform to arrays
ind = range(-window,window+1)
de_r = map(lambda x:de_right.get(x,0), ind)
de_l = map(lambda x:de_left.get(x,0), ind)
#write to files
print "write to files"
fl=open(outprefix+'_count.dat','w')
fl.write('#dist\tX~~\t~~X\n')
for i,j,k in zip(ind,de_r, de_l):
fl.write('{}\t{}\t{}\n'.format(i, j, k))
#write plot
rcParams.update({'font.size': 10})
pp = PdfPages(outprefix+'_plot.pdf')
ind = np.array(ind)
width = 1
pr = plt.bar(ind-0.5, de_r, width, color='r')
pl = plt.bar(ind-0.5, de_l, width, bottom=de_r, color='b')
plt.ylabel("Count")
plt.title("Histogram of counts around cut site")
plt.xticks(ind[::2], rotation="vertical")
plt.legend((pl[0], pr[0]), ("~~X", "X~~"))
plt.gca().set_xlim([-window-1,window+1])
pp.savefig()
pp.close()
def moving_average(a, n=3):
ret = np.cumsum(a, dtype=float)
ret[n:] = ret[n:] - ret[:-n]
return ret[n - 1:] / n
def plot_diagonal_distributions(reads_file, outprefix, ma_window=20,
maxdist=800, de_left=[-2,3], de_right=[0,5]):
rbreaks={}
rejoined={}
des={}
print "process reads"
fl=open(reads_file)
while True:
line=fl.next()
if not line.startswith('#'):
break
nreads=0
try:
while True:
nreads += 1
if nreads % 1000000 == 0:
print nreads
try:
_, n1, sb1, sd1, _, ru1, rd1, n2, sb2, sd2, _, ru2, rd2\
= line.split()
sb1, sd1, ru1, rd1, sb2, sd2, ru2, rd2 = \
map(int, [sb1, sd1, ru1, rd1, sb2, sd2, ru2, rd2])
except ValueError:
print line
raise ValueError("line is not the right format!")
if n1 != n2:
line=fl.next()
continue
#read1 ahead of read2
if sb1 > sb2:
sb1, sd1, ru1, rd1, sb2, sd2, ru2, rd2 = \
sb2, sd2, ru2, rd2, sb1, sd1, ru1, rd1
#direction always -> <-
if not (sd1 == 1 and sd2 == 0):
line=fl.next()
continue
mollen = sb2-sb1
if mollen > maxdist:
line=fl.next()
continue
#DE1
if abs(sb1-ru1) < abs(sb1-rd1):
rc1=ru1
else:
rc1=rd1
pos=sb1-rc1
if pos in de_right:
if not mollen in des:
des[mollen]=0
des[mollen]+=1
line=fl.next()
continue
#DE2
if abs(sb2-ru2) < abs(sb2-rd2):
rc2=ru2
else:
rc2=rd2
pos=sb2-rc2
if pos in de_left:
if not mollen in des:
des[mollen]=0
des[mollen]+=1
line=fl.next()
continue
#random: map on same fragment
if rd1 == rd2:
if not mollen in rbreaks:
rbreaks[mollen]=0
rbreaks[mollen]+=1
line=fl.next()
continue
#rejoined ends
if not mollen in rejoined:
rejoined[mollen]=0
rejoined[mollen]+=1
line=fl.next()
except StopIteration:
pass
print " finished processing {} reads".format(nreads)
#transform to arrays
maxlen = max(max(rejoined),max(des),max(rbreaks))
ind = range(1,maxlen+1)
des = map(lambda x:des.get(x,0), ind)
rbreaks = map(lambda x:rbreaks.get(x,0), ind)
rejoined = map(lambda x:rejoined.get(x,0), ind)
#reweight corner for rejoined
rejoined = map(lambda x: x**.5 * rejoined[x-1]/x, ind)
#write to files
print "write to files"
fl=open(outprefix+'_count.dat','w')
fl.write('#dist\trbreaks\tdes\trejoined\n')
for i,j,k,l in zip(ind,rbreaks,des,rejoined):
fl.write('{}\t{}\t{}\t{}\n'.format(i, j, k, l))
#transform data a bit more
ind, des, rbreaks, rejoined = \
map(lambda x: moving_average(np.array(x), ma_window),
[ind, des, rbreaks, rejoined])
des, rbreaks, rejoined = map(lambda x:x/float(x.sum()),
[des, rbreaks, rejoined])
np.insert(ind,0,0)
np.insert(des,0,0)
np.insert(rbreaks,0,0)
np.insert(rejoined,0,0)
#write plot
pp = PdfPages(outprefix+'_plot.pdf')
rcParams.update({'font.size': 10})
pde = plt.fill_between(ind, des, 0, color='r', alpha=0.5)
prb = plt.fill_between(ind, rbreaks, 0, color='b', alpha=0.5)
prj = plt.fill_between(ind, rejoined, 0, color='y', alpha=0.5)
plt.ylabel("Normalized count")
plt.ylabel("Putative DNA molecule length")
plt.title("Histogram of counts close to the diagonal")
#plt.xticks(ind[::10], rotation="vertical")
plt.legend((prb, pde, prj), ("Random breaks", "Dangling ends",
"Rejoined"))
plt.gca().set_xlim([0,maxlen])
pp.savefig()
pp.close()
def plot_strand_bias_by_distance(fnam, nreads=1000000, valid_pairs=True,
half_step=20, half_len=2000,
full_step=500, full_len=50000, savefig=None):
"""
Classify reads into four categories depending on the strand on which each
of its end is mapped, and plots the proportion of each of these categories
in function of the genomic distance between them.
Only full mapped reads mapped on two diferent restriction fragments (still
same chromosome) are considered.
The four categories are:
- Both read-ends mapped on the same strand (forward)
- Both read-ends mapped on the same strand (reverse)
- Both read-ends mapped on the different strand (facing), like extra-dangling-ends
- Both read-ends mapped on the different strand (opposed), like extra-self-circles
:params fnam: path to tsv file with intersection of mapped ends
:params True valid_pairs: consider only read-ends mapped
on different restriction fragments. If False, considers only read-ends
mapped on the same restriction fragment.
:params 1000000 nreads: number of reads used to plot (if None, all will be used)
:params 20 half_step: binning for the first part of the plot
:params 2000 half_len: maximum distance for the first part of the plot
:params 500 full_step: binning for the second part of the plot
:params 50000 full_len: maximum distance for the second part of the plot
:params None savefig: path to save figure
"""
max_len = 100000
genome_seq = OrderedDict()
pos = 0
fhandler = open(fnam)
for line in fhandler:
if line.startswith('#'):
if line.startswith('# CRM '):
crm, clen = line[6:].split('\t')
genome_seq[crm] = int(clen)
else:
break
pos += len(line)
fhandler.seek(pos)
names = ['<== <== both reverse',
'<== ==> opposed (Extra-self-circles)',
'==> <== facing (Extra-dangling-ends)',
'==> ==> both forward']
dirs = [[0 for i in range(max_len)],
[0 for i in range(max_len)],
[0 for i in range(max_len)],
[0 for i in range(max_len)]]
iterator = (fhandler.next() for _ in xrange(nreads)) if nreads else fhandler
if valid_pairs:
comp_re = lambda x, y: x != y
else:
comp_re = lambda x, y: x == y
for line in iterator:
(crm1, pos1, dir1, len1, re1, _,
crm2, pos2, dir2, len2, re2) = line.strip().split('\t')[1:12]
pos1, pos2 = int(pos1), int(pos2)
if pos2 < pos1:
pos2, pos1 = pos1, pos2
dir2, dir1 = dir1, dir2
len2, len1 = len1, len2
dir1, dir2 = int(dir1), int(dir2)
len1, len2 = int(len1), int(len2)
if dir1 == 0:
pos1 -= len1
if dir2 == 1:
pos2 += len2
diff = pos2 - pos1
# only ligated; same chromsome; bellow max_dist; not multi-contact
if comp_re(re1, re2) and crm1 == crm2 and diff < max_len and len1 == len2:
dir1, dir2 = dir1 * 2, dir2
dirs[dir1 + dir2][diff] += 1
sum_dirs = [0 for i in range(max_len)]
for i in range(max_len):
sum_dir = float(sum(dirs[d][i] for d in range(4)))
for d in range(4):
try:
dirs[d][i] = dirs[d][i] / sum_dir
except ZeroDivisionError:
dirs[d][i] = 0.
sum_dirs[i] = sum_dir
plt.figure(figsize=(14, 9))
if full_step:
axLp = plt.subplot2grid((3, 2), (0, 0), rowspan=2)
axLb = plt.subplot2grid((3, 2), (2, 0), sharex=axLp)
axRp = plt.subplot2grid((3, 2), (0, 1), rowspan=2, sharey=axLp)
axRb = plt.subplot2grid((3, 2), (2, 1), sharex=axRp, sharey=axLb)
else:
axLp = plt.subplot2grid((3, 1), (0, 0), rowspan=2)
axLb = plt.subplot2grid((3, 1), (2, 0), sharex=axLp)
for d in range(4):
axLp.plot([sum(dirs[d][i:i + half_step]) / half_step
for i in range(0, half_len - half_step, half_step)],
alpha=0.7, label=names[d])
axLp.set_ylim(0, 1)
axLp.set_yticks([0, 0.25, 0.5, 0.75, 1])
axLp.set_xlim(0, half_len / half_step)
axLp.set_xticks(axLp.get_xticks()[:-1])
axLp.set_xticklabels([str(int(i)) for i in axLp.get_xticks() * half_step])
axLp.grid()
if full_step:
axLp.spines['right'].set_visible(False)
plt.setp(axLp.get_xticklabels(), visible=False)
axLb.spines['right'].set_visible(False)
axLp.set_ylabel('Proportion of reads in each category')
axLb.bar(range(0, half_len / half_step - 1),
[sum(sum_dirs[i:i + half_step]) / half_step
for i in range(0, half_len - half_step, half_step)],
alpha=0.5, color='k')
axLb.set_ylabel("Log number of reads\nper genomic position")
axLb.set_yscale('log')
axLb.grid()
axLb.set_xlabel('Distance between mapping position of the two ends\n'
'(averaged in windows of 20 nucleotides)')
if full_step:
for d in range(4):
axRp.plot([sum(dirs[d][i:i + full_step]) / full_step
for i in range(half_len, full_len + full_step, full_step)],
alpha=0.7, label=names[d])
axRp.spines['left'].set_visible(False)
axRp.set_xlim(0, full_len / full_step - 2000 / full_step)
axRp.set_xticks(range((10000 - half_step) / full_step, (full_len + full_step) / full_step, 20))
axRp.set_xticklabels([int(i) for i in range(10000, full_len + full_step, full_step * 20)])
plt.setp(axRp.get_xticklabels(), visible=False)
axRp.legend(title='Strand on which each read-end is mapped\n(first read-end is always smaller than second)')
axRp.yaxis.tick_right()
axRp.tick_params(labelleft=False)
axRp.tick_params(labelright=False)
axRp.grid()
axRb.bar(range(0, full_len / full_step - half_len / full_step + 1),
[sum(sum_dirs[i:i + full_step]) / full_step
for i in range(half_len, full_len + full_step, full_step)],
alpha=0.5, color='k')
axRb.set_ylim(0, max(sum_dirs) * 1.1)
axRb.spines['left'].set_visible(False)
axRb.yaxis.tick_right()
axRb.tick_params(labelleft=False)
axRb.tick_params(labelright=False)
axRb.set_xlabel('Distance between mapping position of the two ends\n'
'(averaged in windows of 500 nucleotide)')
axRb.set_yscale('log')
axRb.grid()
# decorate...
d = .015 # how big to make the diagonal lines in axes coordinates
# arguments to pass to plot, just so we don't keep repeating them
kwargs = dict(transform=axLp.transAxes, color='k', clip_on=False)
axLp.plot((1 - d, 1 + d), (1-d, 1+d), **kwargs) # top-left diagonal
axLp.plot((1 - d, 1 + d), (-d, +d), **kwargs) # top-right diagonal
kwargs.update(transform=axRp.transAxes) # switch to the bottom axes
axRp.plot((-d, +d), (1 - d, 1 + d), **kwargs) # bottom-left diagonal
axRp.plot((-d, +d), (-d, +d), **kwargs) # bottom-right diagonal
w = .015
h = .030
# arguments to pass to plot, just so we don't keep repeating them
kwargs = dict(transform=axLb.transAxes, color='k', clip_on=False)
axLb.plot((1 - w, 1 + w), (1 - h, 1 + h), **kwargs) # top-left diagonal
axLb.plot((1 - w, 1 + w), ( - h, + h), **kwargs) # top-right diagonal
kwargs.update(transform=axRb.transAxes) # switch to the bottom axes
axRb.plot((- w, + w), (1 - h, 1 + h), **kwargs) # bottom-left diagonal
axRb.plot((- w, + w), ( - h, + h), **kwargs) # bottom-right diagonal
plt.subplots_adjust(wspace=0.05)
plt.subplots_adjust(hspace=0.1)
else:
axLp.legend(title='Strand on which each read-end is mapped\n(first read-end is always smaller than second)')
if savefig:
tadbit_savefig(savefig)
else:
plt.show()
# For back compatibility
def insert_sizes(fnam, savefig=None, nreads=None, max_size=99.9, axe=None,
show=False, xlog=False, stats=('median', 'perc_max'),
too_large=10000):
"""
Deprecated function, use fragment_size
"""
warn("WARNING: function has been replaced by fragment_size", category=DeprecationWarning,)
return fragment_size(fnam, savefig=savefig, nreads=nreads, max_size=max_size, axe=axe,
show=show, xlog=xlog, stats=stats,
too_large=too_large)
| gpl-3.0 |
google/dl_bounds | dl_bounds/src/data.py | 1 | 5898 | # coding=utf-8
# Copyright 2018 Google LLC
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Dataset retrieval."""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import cPickle
import os
from dl_bounds.src.exp_helpers import flip_labels
from dl_bounds.src.exp_helpers import get_split
import numpy as np
from scipy.io import loadmat
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelBinarizer
from sklearn.preprocessing import StandardScaler
import tensorflow as tf
def get_mnist(data_path, val_size=10000):
ds = tf.contrib.learn.datasets.mnist.read_data_sets(
data_path, one_hot=False, validation_size=val_size, seed=1)
return (ds.train.images, ds.train.labels, ds.validation.images,
ds.validation.labels)
def get_cifar10(data_path):
"""Returns cifar10 dataset.
Args:
data_path: dataset location.
Returns:
tuple (training instances, training labels,
testing instances, testing labels)
Instances of dimension # of instances X dimension.
"""
x_train = np.zeros((50000, 3072))
y_train = np.zeros((50000,), dtype=int)
x_val = np.zeros((10000, 3072))
y_val = np.zeros((10000,), dtype=int)
cur = 0
for batch_index in range(1, 6):
with tf.gfile.Open(
os.path.join(data_path,
"cifar-10-batches-py/data_batch_%d" % batch_index),
"rb") as fo:
batch_data = cPickle.load(fo)
m = batch_data["data"].shape[0]
x_train[cur:cur + m, :] = batch_data["data"].astype(np.float32)
y_train[cur:cur + m] = np.array(batch_data["labels"])
cur += m
assert cur == 50000
with tf.gfile.Open(
os.path.join(data_path, "cifar-10-batches-py/test_batch"), "rb") as fo:
batch_data = cPickle.load(fo)
x_val = batch_data["data"].astype(np.float32)
y_val = np.array(batch_data["labels"])
x_train /= 255.0
x_val /= 255.0
return (x_train, y_train, x_val, y_val)
def get_data(dataset_name, data_path, split_i, split_n, flip_label_ratio=0):
"""Returns a dataset or a given split.
Args:
dataset_name: possible choice: cifar10, mnist, covtype.
data_path: dataset location.
split_i: split index.
split_n: number of examples per split. If -1 -- returns the whole dataset.
flip_label_ratio: randomly flips given amount of labels in the
training and testing sets.
Returns:
tuple (training instances, training labels,
testing instances, testing labels)
Instances of dimension # of instances X dimension.
"""
if dataset_name == "mnist":
(x, y, _, _) = get_mnist(data_path)
# Subsampling valdation set from the training set
# (concerned that val follows a sligtly different distribution)
x_train, x_val, y_train, y_val = train_test_split(
x, y, test_size=0.2, random_state=1)
elif dataset_name == "cifar10":
(x_train, y_train, x_val, y_val) = get_cifar10(data_path)
elif dataset_name == "covtype":
with tf.gfile.Open(os.path.join(data_path, "covtype.mat"), "r") as fh:
mat = loadmat(fh)
x, y = mat["data"].T.todense(), mat["label"].squeeze()
y -= 1
StandardScaler(copy=False, with_mean=True, with_std=True).fit_transform(x)
x_train, x_val, y_train, y_val = train_test_split(
x, y, test_size=0.33, random_state=1)
if split_n > 0: # For negative split_n, return all the data
x_train, y_train = get_split(x_train, y_train, split_i, split_n)
num_classes = len(set(y_train))
if flip_label_ratio > 0:
tf.logging.info("Flipping %f%% of labels in the training set",
flip_label_ratio * 100)
y_train = flip_labels(y_train, flip_label_ratio)
y_val = flip_labels(y_val, flip_label_ratio)
assert (y_train.min() == 0) and (y_val.min() == 0)
lb = LabelBinarizer()
y_train = lb.fit_transform(y_train)
y_val = lb.transform(y_val)
return (x_train, y_train, x_val, y_val, num_classes)
class LocalDatasetProvider(object):
"""Data provider for an in-memory dataset."""
def __init__(self, x, y, limit_size=-1, shuffle_seed=1):
self.x = x
self.y = y
self.index = None
self.rand = None
self.reset_and_reshuffle(shuffle_seed)
self.limit_size(limit_size)
def get_input_dim(self):
return self.x.shape[1]
def get_output_dim(self):
return self.y.shape[1]
def has_more(self):
return self.cur < self.size
def read_next(self, n):
if self.cur <= self.size:
n_read = min(self.size - self.cur, n)
x_mb = self.x[self.index[self.cur:self.cur + n_read], :]
y_mb = self.y[self.index[self.cur:self.cur + n_read], :]
leave_out_indices = np.where(y_mb[:, 0] == -1)[0]
if leave_out_indices:
x_mb = np.delete(x_mb, leave_out_indices, axis=0)
y_mb = np.delete(y_mb, leave_out_indices, axis=0)
n_read = x_mb.shape[0]
self.cur += n_read
return x_mb, y_mb
else:
raise Exception("End-of-dataset.")
def limit_size(self, new_size):
if new_size != -1:
self.size = new_size
else:
self.size = self.x.shape[0]
def reset(self):
self.cur = 0
def reset_and_reshuffle(self, shuffle_seed):
self.cur = 0
self.index = np.arange(self.x.shape[0])
self.rand = np.random.RandomState(shuffle_seed)
self.rand.shuffle(self.index)
| apache-2.0 |
yanchen036/tensorflow | tensorflow/examples/learn/text_classification_character_rnn.py | 38 | 4036 | # Copyright 2016 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Example of recurrent neural networks over characters for DBpedia dataset.
This model is similar to one described in this paper:
"Character-level Convolutional Networks for Text Classification"
http://arxiv.org/abs/1509.01626
and is somewhat alternative to the Lua code from here:
https://github.com/zhangxiangxiao/Crepe
"""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import argparse
import sys
import numpy as np
import pandas
import tensorflow as tf
FLAGS = None
MAX_DOCUMENT_LENGTH = 100
HIDDEN_SIZE = 20
MAX_LABEL = 15
CHARS_FEATURE = 'chars' # Name of the input character feature.
def char_rnn_model(features, labels, mode):
"""Character level recurrent neural network model to predict classes."""
byte_vectors = tf.one_hot(features[CHARS_FEATURE], 256, 1., 0.)
byte_list = tf.unstack(byte_vectors, axis=1)
cell = tf.nn.rnn_cell.GRUCell(HIDDEN_SIZE)
_, encoding = tf.nn.static_rnn(cell, byte_list, dtype=tf.float32)
logits = tf.layers.dense(encoding, MAX_LABEL, activation=None)
predicted_classes = tf.argmax(logits, 1)
if mode == tf.estimator.ModeKeys.PREDICT:
return tf.estimator.EstimatorSpec(
mode=mode,
predictions={
'class': predicted_classes,
'prob': tf.nn.softmax(logits)
})
loss = tf.losses.sparse_softmax_cross_entropy(labels=labels, logits=logits)
if mode == tf.estimator.ModeKeys.TRAIN:
optimizer = tf.train.AdamOptimizer(learning_rate=0.01)
train_op = optimizer.minimize(loss, global_step=tf.train.get_global_step())
return tf.estimator.EstimatorSpec(mode, loss=loss, train_op=train_op)
eval_metric_ops = {
'accuracy': tf.metrics.accuracy(
labels=labels, predictions=predicted_classes)
}
return tf.estimator.EstimatorSpec(
mode=mode, loss=loss, eval_metric_ops=eval_metric_ops)
def main(unused_argv):
# Prepare training and testing data
dbpedia = tf.contrib.learn.datasets.load_dataset(
'dbpedia', test_with_fake_data=FLAGS.test_with_fake_data)
x_train = pandas.DataFrame(dbpedia.train.data)[1]
y_train = pandas.Series(dbpedia.train.target)
x_test = pandas.DataFrame(dbpedia.test.data)[1]
y_test = pandas.Series(dbpedia.test.target)
# Process vocabulary
char_processor = tf.contrib.learn.preprocessing.ByteProcessor(
MAX_DOCUMENT_LENGTH)
x_train = np.array(list(char_processor.fit_transform(x_train)))
x_test = np.array(list(char_processor.transform(x_test)))
# Build model
classifier = tf.estimator.Estimator(model_fn=char_rnn_model)
# Train.
train_input_fn = tf.estimator.inputs.numpy_input_fn(
x={CHARS_FEATURE: x_train},
y=y_train,
batch_size=128,
num_epochs=None,
shuffle=True)
classifier.train(input_fn=train_input_fn, steps=100)
# Eval.
test_input_fn = tf.estimator.inputs.numpy_input_fn(
x={CHARS_FEATURE: x_test},
y=y_test,
num_epochs=1,
shuffle=False)
scores = classifier.evaluate(input_fn=test_input_fn)
print('Accuracy: {0:f}'.format(scores['accuracy']))
if __name__ == '__main__':
parser = argparse.ArgumentParser()
parser.add_argument(
'--test_with_fake_data',
default=False,
help='Test the example code with fake data.',
action='store_true')
FLAGS, unparsed = parser.parse_known_args()
tf.app.run(main=main, argv=[sys.argv[0]] + unparsed)
| apache-2.0 |
timqian/sms-tools | lectures/5-Sinusoidal-model/plots-code/sineModelAnal-flute.py | 24 | 1179 | import numpy as np
import matplotlib.pyplot as plt
import sys, os, time
sys.path.append(os.path.join(os.path.dirname(os.path.realpath(__file__)), '../../../software/models/'))
import stft as STFT
import sineModel as SM
import utilFunctions as UF
(fs, x) = UF.wavread(os.path.join(os.path.dirname(os.path.realpath(__file__)), '../../../sounds/flute-A4.wav'))
w = np.blackman(601)
N = 1024
H = 150
t = -80
minSineDur = .1
maxnSines = 150
mX, pX = STFT.stftAnal(x, fs, w, N, H)
tfreq, tmag, tphase = SM.sineModelAnal(x, fs, w, N, H, t, maxnSines, minSineDur)
plt.figure(1, figsize=(9.5, 5))
maxplotfreq = 5000.0
maxplotbin = int(N*maxplotfreq/fs)
numFrames = int(mX[:,0].size)
frmTime = H*np.arange(numFrames)/float(fs)
binFreq = np.arange(maxplotbin+1)*float(fs)/N
plt.pcolormesh(frmTime, binFreq, np.transpose(mX[:,:maxplotbin+1]))
plt.autoscale(tight=True)
tracks = tfreq*np.less(tfreq, maxplotfreq)
tracks[tracks<=0] = np.nan
plt.plot(frmTime, tracks, color='k', lw=1.5)
plt.autoscale(tight=True)
plt.title('mX + sinusoidal tracks (flute-A4.wav)')
plt.tight_layout()
plt.savefig('sineModelAnal-flute.png')
plt.show() | agpl-3.0 |
nealbob/nealbob.github.io | _site/code/multicore_storage_sim.py | 2 | 2177 | import numpy as np
from matplotlib import pyplot as plt
import time
from multiprocessing import Process
from multiprocessing.queues import Queue
def retry_on_eintr(function, *args, **kw):
while True:
try:
return function(*args, **kw)
except IOError, e:
if e.errno == errno.EINTR:
continue
else:
raise
class RetryQueue(Queue):
"""Queue which will retry if interrupted with EINTR."""
def get(self, block=True, timeout=None):
return retry_on_eintr(Queue.get, self, block, timeout)
def simulate(K, mu, sig, Sbar, T, multi=False, que=0, jobno=0):
np.random.seed(jobno)
S = np.zeros(T+1)
W = np.zeros(T+1)
I = np.zeros(T+1)
S[0] = K
for t in range(T):
W[t] = min(S[t], Sbar)
I[t+1] = max(np.random.normal(mu, sig), 0)
S[t+1] = min(S[t] - W[t] + I[t+1], K)
if multi:
que.put(S)
else:
return S
def multi_sim(CORES=2, T=100):
results = []
ques = [Queue() for i in range(CORES)]
args = [(100, 70, 70, 70, int(T/CORES), True, ques[i], i) for i in range(CORES)]
jobs = [Process(target=simulate, args=(a)) for a in args]
for j in jobs: j.start()
for q in ques: results.append(q.get())
for j in jobs: j.join()
S = np.hstack(results)
return S
"""
### Sample size
T = 1000000
# Single core run ==================================
tic = time.time()
S = simulate(100, 70, 70, 70, T)
toc = time.time()
print 'Single core run time: ' + str(round(toc - tic,3))
plt.plot(S[0:100])
plt.show()
# Multi core run ==================================
tic = time.time()
CORES = 2
results = []
ques = [Queue() for i in range(CORES)]
args = [(100, 70, 70, 70, int(T/CORES), True, ques[i], i) for i in range(CORES)]
jobs = [Process(target=simulate, args=(a)) for a in args]
for j in jobs: j.start()
for q in ques: results.append(q.get())
for j in jobs: j.join()
S = np.hstack(results)
toc = time.time()
print 'Multi-core run time: ' + str(toc - tic)
plt.plot(S[0:100])
plt.show()
print S.shape
plt.scatter(results[0], results[1])
plt.show()
"""
| mit |
echohenry2006/tvb-library | contrib/from_articles/region_deterministic_bnm_wc.py | 5 | 3642 | # -*- coding: utf-8 -*-
"""
What:
Reproduces Figures 23 and 24 of Sanz-Leon P., Knock, S. A., Spiegler, A. and Jirsa V.
Mathematical framework for large-scale brain network modelling in The Virtual Brain.
Neuroimage, 2014, (in review)
Needs:
A working installation of tvb
Run:
python region_deterministic_bnm_wc.py -s True -f True
#Subsequent calls can be made with:
python region_deterministic_bnm_wc.py -f True
.. author:: Paula Sanz-Leon
"""
import numpy
import argparse
from tvb.simulator.lab import *
import matplotlib.pylab as pylab
pylab.rcParams['figure.figsize'] = 19.42, 12 # that's default image size for this interactive session
pylab.rcParams.update({'font.size': 22})
parser = argparse.ArgumentParser(description='Reproduce results of Figure XX presented in Sanz-Leon et al 2014')
parser.add_argument('-s','--sim', help='Run the simulations', default=False)
parser.add_argument('-f','--fig', help='Plot the figures', default=False)
args = vars(parser.parse_args())
speed = 4.0
simulation_length = 512
oscilator = models.WilsonCowan(c_1 = 16., c_2=12., c_3=15., c_4=3, tau_e=8., tau_i=8., a_e=1.3, a_i=2., theta_e=4., theta_i=3.7)
white_matter = connectivity.Connectivity(load_default=True)
white_matter.speed = numpy.array([speed])
gcs = 8
white_matter_coupling = coupling.Linear(a=2**-gcs)
#Initialise an Integrator
heunint = integrators.HeunDeterministic(dt=2**-4)
#Initialise some Monitors with period in physical time
momo = monitors.Raw()
mama = monitors.TemporalAverage(period=2**-2)
#Bundle them
what_to_watch = (momo, mama)
#Initialise a Simulator -- Model, Connectivity, Integrator, and Monitors.
sim = simulator.Simulator(model = oscilator, connectivity = white_matter,
coupling = white_matter_coupling,
integrator = heunint, monitors = what_to_watch)
sim.configure()
LOG.info("Starting simulation...")
#Perform the simulation
raw_data = []
raw_time = []
tavg_data = []
tavg_time = []
for raw, tavg in sim(simulation_length=simulation_length):
if not raw is None:
raw_time.append(raw[0])
raw_data.append(raw[1])
if not tavg is None:
tavg_time.append(tavg[0])
tavg_data.append(tavg[1])
LOG.info("Finished simulation.")
#Make the lists numpy.arrays for easier use.
RAW = numpy.array(raw_data)
TAVG = numpy.array(tavg_data)
# <codecell>
numpy.save('region_deterministic_bnm_article_wc_raw.npy', RAW)
numpy.save('region_deterministic_bnm_article_wc_rawtime.npy', raw_time)
numpy.save('region_deterministic_bnm_article_wc_tavg.npy', TAVG)
numpy.save('region_deterministic_bnm_article_wc_tavgtime.npy', tavg_time)
if args['fig']:
RAW = numpy.load('region_deterministic_bnm_article_wc_raw.npy')
raw_time = numpy.load('region_deterministic_bnm_article_wc_rawtime.npy')
#Plot temporally averaged time series
figure(1)
subplot(1, 2, 1)
plot(raw_time, RAW[:, 0, :, 0], 'k', alpha=0.042, linewidth=3)
plot(raw_time, RAW[:, 1, :, 0], 'r', alpha=0.042, linewidth=3)
plot(raw_time, RAW[:, 0, :, 0].mean(axis=1), 'k', linewidth=3)
plot(raw_time, RAW[:, 1, :, 0].mean(axis=1), 'r', linewidth=3)
xlabel('time[ms]')
#ylim([-25, 5])
xlim([0, sim.simulation_length])
subplot(1, 2, 2)
plot(RAW[:, 0, :, 0], RAW[:, 1, :, 0], alpha=0.042)
plot(RAW[:, 0, :, 0].mean(axis=1), RAW[:, 1, :, 0].mean(axis=1), alpha=1.)
plot(RAW[0, 0, :, 0], RAW[0, 1, :, 0], 'bo', alpha=0.15)
xlabel(r'$E$')
ylabel(r'$I$')
show()
fig_name = 'wc_default_speed_' + str(int(white_matter.speed)) + '_gcs_2**-' + str(gcs) + '.pdf'
savefig(fig_name)
###EoF### | gpl-2.0 |
HaebinShin/tensorflow | tensorflow/examples/skflow/hdf5_classification.py | 5 | 2006 | # Copyright 2016 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
"""Example of DNNClassifier for Iris plant dataset, h5 format."""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
from sklearn import cross_validation
from sklearn import metrics
from tensorflow.contrib import learn
import h5py # pylint: disable=g-bad-import-order
# Load dataset.
iris = learn.datasets.load_dataset('iris')
x_train, x_test, y_train, y_test = cross_validation.train_test_split(
iris.data, iris.target, test_size=0.2, random_state=42)
# Note that we are saving and load iris data as h5 format as a simple
# demonstration here.
h5f = h5py.File('test_hdf5.h5', 'w')
h5f.create_dataset('X_train', data=x_train)
h5f.create_dataset('X_test', data=x_test)
h5f.create_dataset('y_train', data=y_train)
h5f.create_dataset('y_test', data=y_test)
h5f.close()
h5f = h5py.File('test_hdf5.h5', 'r')
x_train = h5f['X_train']
x_test = h5f['X_test']
y_train = h5f['y_train']
y_test = h5f['y_test']
# Build 3 layer DNN with 10, 20, 10 units respectively.
feature_columns = learn.infer_real_valued_columns_from_input(x_train)
classifier = learn.TensorFlowDNNClassifier(
feature_columns=feature_columns, hidden_units=[10, 20, 10], n_classes=3,
steps=200)
# Fit and predict.
classifier.fit(x_train, y_train)
score = metrics.accuracy_score(y_test, classifier.predict(x_test))
print('Accuracy: {0:f}'.format(score))
| apache-2.0 |
grantvk/aima-python | submissions/Sery/myNN.py | 13 | 3375 | from sklearn.neural_network import MLPClassifier
import traceback
from submissions.Sery import aids
class DataFrame:
data = []
feature_names = []
target = []
target_names = []
aidsECHP = DataFrame()
aidsECHP.data = []
target_data = []
list_of_report = aids.get_reports()
for record in list_of_report:
try:
prevalence = float(record['Data']["HIV Prevalence"]["Adults"])
target_data.append(prevalence)
year = int(record['Year'])
living = int(record['Data']["People Living with HIV"]["Adults"])
new = int(record['Data']["New HIV Infections"]["Adults"])
deaths = int(record['Data']["AIDS-Related Deaths"]["Adults"])
aidsECHP.data.append([year, living, new, deaths])
except:
traceback.print_exc()
aidsECHP.feature_names = [
'Year',
'People Living with HIV',
'New HIV Infections',
'AIDS-Related Deaths',
]
'''
Build the target list,
one entry for each row in the input frame.
The Naive Bayesian network is a classifier,
i.e. it sorts data points into bins.
The best it can do to estimate a continuous variable
is to break the domain into segments, and predict
the segment into which the variable's value will fall.
In this example, I'm breaking Trump's % into two
arbitrary segments.
'''
aidsECHP.target = []
def aidsTarget(percentage):
if percentage > 6:
return 1
return 0
for pre in target_data:
# choose the target
tt = aidsTarget(pre)
aidsECHP.target.append(tt)
aidsECHP.target_names = [
'HIV Prevalence <= 6%',
'HIV Prevalence > 6%',
]
'''
Make a customn classifier,
'''
mlpc = MLPClassifier(
hidden_layer_sizes = (120,),
activation = 'relu',
solver='sgd', # 'adam',
alpha = 0.00001,
# batch_size='auto',
learning_rate = 'adaptive', # 'constant',
# power_t = 0.5,
max_iter = 1200, # 200,
shuffle = True,
# random_state = None,
# tol = 1e-4,
# verbose = False,
# warm_start = False,
# momentum = 0.9,
# nesterovs_momentum = True,
# early_stopping = False,
# validation_fraction = 0.1,
# beta_1 = 0.9,
# beta_2 = 0.999,
# epsilon = 1e-8,
)
'''
Try scaling the data.
'''
aidsScaled = DataFrame()
def setupScales(grid):
global min, max
min = list(grid[0])
max = list(grid[0])
for row in range(1, len(grid)):
for col in range(len(grid[row])):
cell = grid[row][col]
if cell < min[col]:
min[col] = cell
if cell > max[col]:
max[col] = cell
def scaleGrid(grid):
newGrid = []
for row in range(len(grid)):
newRow = []
for col in range(len(grid[row])):
try:
cell = grid[row][col]
scaled = (cell - min[col]) \
/ (max[col] - min[col])
newRow.append(scaled)
except:
pass
newGrid.append(newRow)
return newGrid
setupScales(aidsECHP.data)
aidsScaled.data = scaleGrid(aidsECHP.data)
aidsScaled.feature_names = aidsECHP.feature_names
aidsScaled.target = aidsECHP.target
aidsScaled.target_names = aidsECHP.target_names
Examples = {
'AidsDefault': {
'frame': aidsECHP,
},
'AidsSGD': {
'frame': aidsECHP,
'mlpc': mlpc
},
'AidsScaled': {
'frame': aidsScaled,
},
} | mit |
fyffyt/scikit-learn | examples/ensemble/plot_adaboost_regression.py | 311 | 1529 | """
======================================
Decision Tree Regression with AdaBoost
======================================
A decision tree is boosted using the AdaBoost.R2 [1] algorithm on a 1D
sinusoidal dataset with a small amount of Gaussian noise.
299 boosts (300 decision trees) is compared with a single decision tree
regressor. As the number of boosts is increased the regressor can fit more
detail.
.. [1] H. Drucker, "Improving Regressors using Boosting Techniques", 1997.
"""
print(__doc__)
# Author: Noel Dawe <noel.dawe@gmail.com>
#
# License: BSD 3 clause
# importing necessary libraries
import numpy as np
import matplotlib.pyplot as plt
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import AdaBoostRegressor
# Create the dataset
rng = np.random.RandomState(1)
X = np.linspace(0, 6, 100)[:, np.newaxis]
y = np.sin(X).ravel() + np.sin(6 * X).ravel() + rng.normal(0, 0.1, X.shape[0])
# Fit regression model
regr_1 = DecisionTreeRegressor(max_depth=4)
regr_2 = AdaBoostRegressor(DecisionTreeRegressor(max_depth=4),
n_estimators=300, random_state=rng)
regr_1.fit(X, y)
regr_2.fit(X, y)
# Predict
y_1 = regr_1.predict(X)
y_2 = regr_2.predict(X)
# Plot the results
plt.figure()
plt.scatter(X, y, c="k", label="training samples")
plt.plot(X, y_1, c="g", label="n_estimators=1", linewidth=2)
plt.plot(X, y_2, c="r", label="n_estimators=300", linewidth=2)
plt.xlabel("data")
plt.ylabel("target")
plt.title("Boosted Decision Tree Regression")
plt.legend()
plt.show()
| bsd-3-clause |
waynenilsen/statsmodels | statsmodels/tsa/base/tests/test_base.py | 27 | 2106 | import numpy as np
from pandas import Series
from pandas import date_range
from statsmodels.tsa.base.tsa_model import TimeSeriesModel
import numpy.testing as npt
from statsmodels.tools.testing import assert_equal
def test_pandas_nodates_index():
from statsmodels.datasets import sunspots
y = sunspots.load_pandas().data.SUNACTIVITY
npt.assert_raises(ValueError, TimeSeriesModel, y)
def test_predict_freq():
# test that predicted dates have same frequency
x = np.arange(1,36.)
# there's a bug in pandas up to 0.10.2 for YearBegin
#dates = date_range("1972-4-1", "2007-4-1", freq="AS-APR")
dates = date_range("1972-4-30", "2006-4-30", freq="A-APR")
series = Series(x, index=dates)
model = TimeSeriesModel(series)
#npt.assert_(model.data.freq == "AS-APR")
npt.assert_(model.data.freq == "A-APR")
start = model._get_predict_start("2006-4-30")
end = model._get_predict_end("2016-4-30")
model._make_predict_dates()
predict_dates = model.data.predict_dates
#expected_dates = date_range("2006-12-31", "2016-12-31",
# freq="AS-APR")
expected_dates = date_range("2006-4-30", "2016-4-30", freq="A-APR")
assert_equal(predict_dates, expected_dates)
#ptesting.assert_series_equal(predict_dates, expected_dates)
def test_keyerror_start_date():
x = np.arange(1,36.)
from pandas import date_range
# there's a bug in pandas up to 0.10.2 for YearBegin
#dates = date_range("1972-4-1", "2007-4-1", freq="AS-APR")
dates = date_range("1972-4-30", "2006-4-30", freq="A-APR")
series = Series(x, index=dates)
model = TimeSeriesModel(series)
npt.assert_raises(ValueError, model._get_predict_start, "1970-4-30")
def test_period_index():
# test 1285
from pandas import PeriodIndex, TimeSeries
dates = PeriodIndex(start="1/1/1990", periods=20, freq="M")
x = np.arange(1, 21.)
model = TimeSeriesModel(Series(x, index=dates))
npt.assert_(model.data.freq == "M")
model = TimeSeriesModel(TimeSeries(x, index=dates))
npt.assert_(model.data.freq == "M")
| bsd-3-clause |
RayMick/scikit-learn | examples/semi_supervised/plot_label_propagation_digits.py | 268 | 2723 | """
===================================================
Label Propagation digits: Demonstrating performance
===================================================
This example demonstrates the power of semisupervised learning by
training a Label Spreading model to classify handwritten digits
with sets of very few labels.
The handwritten digit dataset has 1797 total points. The model will
be trained using all points, but only 30 will be labeled. Results
in the form of a confusion matrix and a series of metrics over each
class will be very good.
At the end, the top 10 most uncertain predictions will be shown.
"""
print(__doc__)
# Authors: Clay Woolam <clay@woolam.org>
# Licence: BSD
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
from sklearn import datasets
from sklearn.semi_supervised import label_propagation
from sklearn.metrics import confusion_matrix, classification_report
digits = datasets.load_digits()
rng = np.random.RandomState(0)
indices = np.arange(len(digits.data))
rng.shuffle(indices)
X = digits.data[indices[:330]]
y = digits.target[indices[:330]]
images = digits.images[indices[:330]]
n_total_samples = len(y)
n_labeled_points = 30
indices = np.arange(n_total_samples)
unlabeled_set = indices[n_labeled_points:]
# shuffle everything around
y_train = np.copy(y)
y_train[unlabeled_set] = -1
###############################################################################
# Learn with LabelSpreading
lp_model = label_propagation.LabelSpreading(gamma=0.25, max_iter=5)
lp_model.fit(X, y_train)
predicted_labels = lp_model.transduction_[unlabeled_set]
true_labels = y[unlabeled_set]
cm = confusion_matrix(true_labels, predicted_labels, labels=lp_model.classes_)
print("Label Spreading model: %d labeled & %d unlabeled points (%d total)" %
(n_labeled_points, n_total_samples - n_labeled_points, n_total_samples))
print(classification_report(true_labels, predicted_labels))
print("Confusion matrix")
print(cm)
# calculate uncertainty values for each transduced distribution
pred_entropies = stats.distributions.entropy(lp_model.label_distributions_.T)
# pick the top 10 most uncertain labels
uncertainty_index = np.argsort(pred_entropies)[-10:]
###############################################################################
# plot
f = plt.figure(figsize=(7, 5))
for index, image_index in enumerate(uncertainty_index):
image = images[image_index]
sub = f.add_subplot(2, 5, index + 1)
sub.imshow(image, cmap=plt.cm.gray_r)
plt.xticks([])
plt.yticks([])
sub.set_title('predict: %i\ntrue: %i' % (
lp_model.transduction_[image_index], y[image_index]))
f.suptitle('Learning with small amount of labeled data')
plt.show()
| bsd-3-clause |
Stonelinks/jsbsim | tests/CheckOutputRate.py | 2 | 6059 | # CheckOutputRate.py
#
# A regression test on the output features that allow to set the output rate
# including enabling/disabling the output.
#
# Copyright (c) 2015 Bertrand Coconnier
#
# This program is free software; you can redistribute it and/or modify it under
# the terms of the GNU General Public License as published by the Free Software
# Foundation; either version 3 of the License, or (at your option) any later
# version.
#
# This program is distributed in the hope that it will be useful, but WITHOUT
# ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS
# FOR A PARTICULAR PURPOSE. See the GNU General Public License for more
# details.
#
# You should have received a copy of the GNU General Public License along with
# this program; if not, see <http://www.gnu.org/licenses/>
#
import string
import xml.etree.ElementTree as et
import pandas as pd
from JSBSim_utils import JSBSimTestCase, CreateFDM, append_xml, RunTest
class CheckOutputRate(JSBSimTestCase):
def setUp(self):
JSBSimTestCase.setUp(self)
self.fdm = CreateFDM(self.sandbox)
self.script_path = self.sandbox.path_to_jsbsim_file('scripts',
'c1722.xml')
# Read the time step 'dt' from the script file
self.tree = et.parse(self.script_path)
root = self.tree.getroot()
use_tag = root.find('use')
aircraft_name = use_tag.attrib['aircraft']
self.run_tag = root.find('run')
self.dt = float(self.run_tag.attrib['dt'])
# Read the date at which the trim will be run
for event in root.findall('run/event'):
if event.attrib['name'] == 'Trim':
cond_tag = event.find('condition')
self.trim_date = float(string.split(cond_tag.text)[-1])
break
# Read the output rate and the output file from the aircraft file
aircraft_path = self.sandbox.path_to_jsbsim_file('aircraft', aircraft_name,
append_xml(aircraft_name))
tree = et.parse(aircraft_path)
output_tag = tree.getroot().find('output')
self.output_file = output_tag.attrib['name']
self.rateHz = float(output_tag.attrib['rate'])
self.rate = int(1.0 / (self.rateHz * self.dt))
def tearDown(self):
del self.fdm
JSBSimTestCase.tearDown(self)
def testOutputRate(self):
self.fdm.load_script(self.script_path)
# Check that the output is enabled by default
self.assertEqual(self.fdm["simulation/output/enabled"], 1.0)
# Check that the rate is consistent with the values extracted from the
# script and the aircraft definition
self.assertAlmostEqual(self.fdm["simulation/output/log_rate_hz"],
self.rateHz, delta=1E-5)
self.fdm.run_ic()
for i in xrange(self.rate):
self.fdm.run()
output = pd.read_csv(self.output_file)
# According to the settings, the output file must contain 2 lines in
# addition to the headers :
# 1. The initial conditions
# 2. The output after 'rate' iterations
self.assertEqual(output['Time'].iget(0), 0.0)
self.assertEqual(output['Time'].iget(1), self.rate * self.dt)
self.assertEqual(output['Time'].iget(1),
self.fdm["simulation/sim-time-sec"])
def testDisablingOutput(self):
self.fdm.load_script(self.script_path)
# Disables the output during the initialization
self.fdm["simulation/output/enabled"] = 0.0
self.fdm.run_ic()
self.fdm["simulation/output/enabled"] = 1.0
for i in xrange(self.rate):
self.fdm.run()
output = pd.read_csv(self.output_file)
# According to the settings, the output file must contain 1 line in
# addition to the headers :
# 1. The output after 'rate' iterations
self.assertEqual(output['Time'].iget(0),
self.fdm["simulation/sim-time-sec"])
def testTrimRestoresOutputSettings(self):
self.fdm.load_script(self.script_path)
# Disables the output during the initialization
self.fdm["simulation/output/enabled"] = 0.0
self.fdm.run_ic()
# Check that the output remains disabled even after the trim is
# executed
while self.fdm["simulation/sim-time-sec"] < self.trim_date + 2.0*self.dt:
self.fdm.run()
self.assertEqual(self.fdm["simulation/output/enabled"], 0.0)
# Re-enable the output and check that the output rate is unaffected by
# the previous operations
self.fdm["simulation/output/enabled"] = 1.0
frame = int(self.fdm["simulation/frame"])
for i in xrange(self.rate):
self.fdm.run()
output = pd.read_csv(self.output_file)
# The frame at which the data is logged must be the next multiple of
# the output rate
self.assertEqual(int(output['Time'].iget(0)/self.dt),
(1 + frame/self.rate)*self.rate)
def testDisablingOutputInScript(self):
property = et.SubElement(self.run_tag, 'property')
property.text = 'simulation/output/enabled'
property.attrib['value'] = "0.0"
self.tree.write('c1722_0.xml')
self.fdm.load_script('c1722_0.xml')
# Check that the output is disabled
self.assertEqual(self.fdm["simulation/output/enabled"], 0.0)
self.fdm.run_ic()
self.fdm["simulation/output/enabled"] = 1.0
for i in xrange(self.rate):
self.fdm.run()
output = pd.read_csv(self.output_file)
# According to the settings, the output file must contain 1 line in
# addition to the headers :
# 1. The output after 'rate' iterations
self.assertEqual(output['Time'].iget(0),
self.fdm["simulation/sim-time-sec"])
RunTest(CheckOutputRate)
| lgpl-2.1 |
goldmedal/spark | python/pyspark/sql/tests/test_pandas_udf.py | 5 | 10122 | #
# Licensed to the Apache Software Foundation (ASF) under one or more
# contributor license agreements. See the NOTICE file distributed with
# this work for additional information regarding copyright ownership.
# The ASF licenses this file to You under the Apache License, Version 2.0
# (the "License"); you may not use this file except in compliance with
# the License. You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
#
import unittest
from pyspark.sql.functions import udf, pandas_udf, PandasUDFType
from pyspark.sql.types import *
from pyspark.sql.utils import ParseException
from pyspark.rdd import PythonEvalType
from pyspark.testing.sqlutils import ReusedSQLTestCase, have_pandas, have_pyarrow, \
pandas_requirement_message, pyarrow_requirement_message
from pyspark.testing.utils import QuietTest
from py4j.protocol import Py4JJavaError
@unittest.skipIf(
not have_pandas or not have_pyarrow,
pandas_requirement_message or pyarrow_requirement_message)
class PandasUDFTests(ReusedSQLTestCase):
def test_pandas_udf_basic(self):
udf = pandas_udf(lambda x: x, DoubleType())
self.assertEqual(udf.returnType, DoubleType())
self.assertEqual(udf.evalType, PythonEvalType.SQL_SCALAR_PANDAS_UDF)
udf = pandas_udf(lambda x: x, DoubleType(), PandasUDFType.SCALAR)
self.assertEqual(udf.returnType, DoubleType())
self.assertEqual(udf.evalType, PythonEvalType.SQL_SCALAR_PANDAS_UDF)
udf = pandas_udf(lambda x: x, 'double', PandasUDFType.SCALAR)
self.assertEqual(udf.returnType, DoubleType())
self.assertEqual(udf.evalType, PythonEvalType.SQL_SCALAR_PANDAS_UDF)
udf = pandas_udf(lambda x: x, StructType([StructField("v", DoubleType())]),
PandasUDFType.GROUPED_MAP)
self.assertEqual(udf.returnType, StructType([StructField("v", DoubleType())]))
self.assertEqual(udf.evalType, PythonEvalType.SQL_GROUPED_MAP_PANDAS_UDF)
udf = pandas_udf(lambda x: x, 'v double', PandasUDFType.GROUPED_MAP)
self.assertEqual(udf.returnType, StructType([StructField("v", DoubleType())]))
self.assertEqual(udf.evalType, PythonEvalType.SQL_GROUPED_MAP_PANDAS_UDF)
udf = pandas_udf(lambda x: x, 'v double',
functionType=PandasUDFType.GROUPED_MAP)
self.assertEqual(udf.returnType, StructType([StructField("v", DoubleType())]))
self.assertEqual(udf.evalType, PythonEvalType.SQL_GROUPED_MAP_PANDAS_UDF)
udf = pandas_udf(lambda x: x, returnType='v double',
functionType=PandasUDFType.GROUPED_MAP)
self.assertEqual(udf.returnType, StructType([StructField("v", DoubleType())]))
self.assertEqual(udf.evalType, PythonEvalType.SQL_GROUPED_MAP_PANDAS_UDF)
def test_pandas_udf_decorator(self):
@pandas_udf(DoubleType())
def foo(x):
return x
self.assertEqual(foo.returnType, DoubleType())
self.assertEqual(foo.evalType, PythonEvalType.SQL_SCALAR_PANDAS_UDF)
@pandas_udf(returnType=DoubleType())
def foo(x):
return x
self.assertEqual(foo.returnType, DoubleType())
self.assertEqual(foo.evalType, PythonEvalType.SQL_SCALAR_PANDAS_UDF)
schema = StructType([StructField("v", DoubleType())])
@pandas_udf(schema, PandasUDFType.GROUPED_MAP)
def foo(x):
return x
self.assertEqual(foo.returnType, schema)
self.assertEqual(foo.evalType, PythonEvalType.SQL_GROUPED_MAP_PANDAS_UDF)
@pandas_udf('v double', PandasUDFType.GROUPED_MAP)
def foo(x):
return x
self.assertEqual(foo.returnType, schema)
self.assertEqual(foo.evalType, PythonEvalType.SQL_GROUPED_MAP_PANDAS_UDF)
@pandas_udf(schema, functionType=PandasUDFType.GROUPED_MAP)
def foo(x):
return x
self.assertEqual(foo.returnType, schema)
self.assertEqual(foo.evalType, PythonEvalType.SQL_GROUPED_MAP_PANDAS_UDF)
@pandas_udf(returnType='double', functionType=PandasUDFType.SCALAR)
def foo(x):
return x
self.assertEqual(foo.returnType, DoubleType())
self.assertEqual(foo.evalType, PythonEvalType.SQL_SCALAR_PANDAS_UDF)
@pandas_udf(returnType=schema, functionType=PandasUDFType.GROUPED_MAP)
def foo(x):
return x
self.assertEqual(foo.returnType, schema)
self.assertEqual(foo.evalType, PythonEvalType.SQL_GROUPED_MAP_PANDAS_UDF)
def test_udf_wrong_arg(self):
with QuietTest(self.sc):
with self.assertRaises(ParseException):
@pandas_udf('blah')
def foo(x):
return x
with self.assertRaisesRegexp(ValueError, 'Invalid return type.*None'):
@pandas_udf(functionType=PandasUDFType.SCALAR)
def foo(x):
return x
with self.assertRaisesRegexp(ValueError, 'Invalid function'):
@pandas_udf('double', 100)
def foo(x):
return x
with self.assertRaisesRegexp(ValueError, '0-arg pandas_udfs.*not.*supported'):
pandas_udf(lambda: 1, LongType(), PandasUDFType.SCALAR)
with self.assertRaisesRegexp(ValueError, '0-arg pandas_udfs.*not.*supported'):
@pandas_udf(LongType(), PandasUDFType.SCALAR)
def zero_with_type():
return 1
with self.assertRaisesRegexp(TypeError, 'Invalid return type'):
@pandas_udf(returnType=PandasUDFType.GROUPED_MAP)
def foo(df):
return df
with self.assertRaisesRegexp(TypeError, 'Invalid return type'):
@pandas_udf(returnType='double', functionType=PandasUDFType.GROUPED_MAP)
def foo(df):
return df
with self.assertRaisesRegexp(ValueError, 'Invalid function'):
@pandas_udf(returnType='k int, v double', functionType=PandasUDFType.GROUPED_MAP)
def foo(k, v, w):
return k
def test_stopiteration_in_udf(self):
def foo(x):
raise StopIteration()
def foofoo(x, y):
raise StopIteration()
exc_message = "Caught StopIteration thrown from user's code; failing the task"
df = self.spark.range(0, 100)
# plain udf (test for SPARK-23754)
self.assertRaisesRegexp(
Py4JJavaError,
exc_message,
df.withColumn('v', udf(foo)('id')).collect
)
# pandas scalar udf
self.assertRaisesRegexp(
Py4JJavaError,
exc_message,
df.withColumn(
'v', pandas_udf(foo, 'double', PandasUDFType.SCALAR)('id')
).collect
)
# pandas grouped map
self.assertRaisesRegexp(
Py4JJavaError,
exc_message,
df.groupBy('id').apply(
pandas_udf(foo, df.schema, PandasUDFType.GROUPED_MAP)
).collect
)
self.assertRaisesRegexp(
Py4JJavaError,
exc_message,
df.groupBy('id').apply(
pandas_udf(foofoo, df.schema, PandasUDFType.GROUPED_MAP)
).collect
)
# pandas grouped agg
self.assertRaisesRegexp(
Py4JJavaError,
exc_message,
df.groupBy('id').agg(
pandas_udf(foo, 'double', PandasUDFType.GROUPED_AGG)('id')
).collect
)
def test_pandas_udf_detect_unsafe_type_conversion(self):
import pandas as pd
import numpy as np
values = [1.0] * 3
pdf = pd.DataFrame({'A': values})
df = self.spark.createDataFrame(pdf).repartition(1)
@pandas_udf(returnType="int")
def udf(column):
return pd.Series(np.linspace(0, 1, len(column)))
# Since 0.11.0, PyArrow supports the feature to raise an error for unsafe cast.
with self.sql_conf({
"spark.sql.execution.pandas.convertToArrowArraySafely": True}):
with self.assertRaisesRegexp(Exception,
"Exception thrown when converting pandas.Series"):
df.select(['A']).withColumn('udf', udf('A')).collect()
# Disabling Arrow safe type check.
with self.sql_conf({
"spark.sql.execution.pandas.convertToArrowArraySafely": False}):
df.select(['A']).withColumn('udf', udf('A')).collect()
def test_pandas_udf_arrow_overflow(self):
import pandas as pd
df = self.spark.range(0, 1)
@pandas_udf(returnType="byte")
def udf(column):
return pd.Series([128] * len(column))
# When enabling safe type check, Arrow 0.11.0+ disallows overflow cast.
with self.sql_conf({
"spark.sql.execution.pandas.convertToArrowArraySafely": True}):
with self.assertRaisesRegexp(Exception,
"Exception thrown when converting pandas.Series"):
df.withColumn('udf', udf('id')).collect()
# Disabling safe type check, let Arrow do the cast anyway.
with self.sql_conf({"spark.sql.execution.pandas.convertToArrowArraySafely": False}):
df.withColumn('udf', udf('id')).collect()
if __name__ == "__main__":
from pyspark.sql.tests.test_pandas_udf import *
try:
import xmlrunner
testRunner = xmlrunner.XMLTestRunner(output='target/test-reports', verbosity=2)
except ImportError:
testRunner = None
unittest.main(testRunner=testRunner, verbosity=2)
| apache-2.0 |
PatrickOReilly/scikit-learn | sklearn/feature_selection/__init__.py | 140 | 1302 | """
The :mod:`sklearn.feature_selection` module implements feature selection
algorithms. It currently includes univariate filter selection methods and the
recursive feature elimination algorithm.
"""
from .univariate_selection import chi2
from .univariate_selection import f_classif
from .univariate_selection import f_oneway
from .univariate_selection import f_regression
from .univariate_selection import SelectPercentile
from .univariate_selection import SelectKBest
from .univariate_selection import SelectFpr
from .univariate_selection import SelectFdr
from .univariate_selection import SelectFwe
from .univariate_selection import GenericUnivariateSelect
from .variance_threshold import VarianceThreshold
from .rfe import RFE
from .rfe import RFECV
from .from_model import SelectFromModel
from .mutual_info_ import mutual_info_regression, mutual_info_classif
__all__ = ['GenericUnivariateSelect',
'RFE',
'RFECV',
'SelectFdr',
'SelectFpr',
'SelectFwe',
'SelectKBest',
'SelectFromModel',
'SelectPercentile',
'VarianceThreshold',
'chi2',
'f_classif',
'f_oneway',
'f_regression',
'mutual_info_classif',
'mutual_info_regression']
| bsd-3-clause |
turi-code/SFrame | oss_src/unity/python/sframe/test/util.py | 5 | 4772 | '''
Copyright (C) 2016 Turi
All rights reserved.
This software may be modified and distributed under the terms
of the BSD license. See the LICENSE file for details.
'''
import random
import tempfile
import shutil
import math
import string
import numpy as np
from pandas.util.testing import assert_frame_equal
from .. import SArray
class SFrameComparer():
"""
Helper class for comparing sframe and sarrays
Adapted from test_sframe.py
"""
def _assert_sgraph_equal(self, sg1, sg2):
self._assert_sframe_equal(sg1.vertices, sg2.vertices)
self._assert_sframe_equal(sg1.edges, sg2.edges)
def _assert_sframe_equal(self, sf1, sf2):
assert sf1.num_rows() == sf2.num_rows()
assert sf1.num_cols() == sf2.num_cols()
assert set(sf1.column_names()) == set(sf2.column_names())
assert_frame_equal(sf1.to_dataframe(), sf2.to_dataframe())
def _assert_sarray_equal(self, sa1, sa2):
l1 = list(sa1)
l2 = list(sa2)
assert len(l1) == len(l2)
for i in range(len(l1)):
v1 = l1[i]
v2 = l2[i]
if v1 == None:
assert v2 == None
else:
if type(v1) == dict:
assert len(v1) == len(v2)
for key in v1:
assert key in v1
assert v1[key] == v2[key]
elif (hasattr(v1, "__iter__")):
assert len(v1) == len(v2)
for j in range(len(v1)):
t1 = v1[j]; t2 = v2[j]
if (type(t1) == float):
if (math.isnan(t1)):
assert math.isnan(t2)
else:
assert t1 == t2
else:
assert t1 == t2
else:
assert v1 == v2
class SubstringMatcher():
"""
Helper class for testing substring matching
Code adapted from http://www.michaelpollmeier.com/python-mock-how-to-assert-a-substring-of-logger-output/
"""
def __init__(self, containing):
self.containing = containing.lower()
def __eq__(self, other):
return other.lower().find(self.containing) > -1
def __unicode__(self):
return 'a string containing "%s"' % self.containing
def __str__(self):
return unicode(self).encode('utf-8')
__repr__ = __unicode__
class TempDirectory():
name = None
def __init__(self):
self.name = tempfile.mkdtemp()
def __enter__(self):
return self.name
def __exit__(self, type, value, traceback):
if self.name != None:
shutil.rmtree(self.name)
def uniform_string_column(n, word_length, alphabet_size, missingness=0.):
"""
Return an SArray of strings constructed uniformly randomly from the first
'num_letters' of the lower case alphabet.
Parameters
----------
n : int
Number of entries in the output SArray.
word_length : int
Number of characters in each string.
alphabet_size : int
Number of characters in the alphabet.
missingness : float, optional
Probability that a given entry in the output is missing.
Returns
-------
out : SArray
One string "word" in each entry of the output SArray.
"""
result = []
letters = list(string.ascii_letters[:alphabet_size])
for i in range(n):
missing_flag = random.random()
if missing_flag < missingness:
result.append(None)
else:
word = []
for j in range(word_length):
word.append(np.random.choice(letters))
result.append(''.join(word))
return SArray(result)
def uniform_numeric_column(n, col_type=float, range=(0, 1), missingness=0.):
"""
Return an SArray of uniformly random numeric values.
Parameters
----------
n : int
Number of entries in the output SArray.
col_type : type, optional
Type of the output SArray. Default is floats.
range : tuple[int, int], optional
Minimum and maximum of the uniform distribution from which values are
chosen.
missingness : float, optional
Probability that a given entry in the output is missing.
Returns
-------
out : SArray
"""
if col_type == int:
v = np.random.randint(low=range[0], high=range[1], size=n).astype(float)
else:
v = np.random.rand(n)
v = v * (range[1] - range[0]) + range[0]
idx_na = np.random.rand(n) < missingness
v[idx_na] = None
v = np.where(np.isnan(v), None, v)
return SArray(v, dtype=col_type)
| bsd-3-clause |
ODM2/ODMToolsPython | odmtools/gui/pnlPlot.py | 1 | 7003 | #Boa:FramePanel:Panel1
import wx
from wx.lib.pubsub import pub as Publisher
try:
from agw import flatnotebook as fnb
except ImportError: # if it's not there locally, try the wxPython lib.
import wx.lib.agw.flatnotebook as fnb
import matplotlib
matplotlib.use('WXAgg')
import plotTimeSeries
import plotSummary
import plotHistogram
import plotBoxWhisker
import plotProbability
from odmtools.controller.logicPlotOptions import SeriesPlotInfo
import logging
# from odmtools.common.logger import LoggerTool
#
# tool = LoggerTool()
# logger = tool.setupLogger(__name__, __name__ + '.log', 'w', logging.DEBUG)
logger =logging.getLogger('main')
[wxID_PANEL1, wxID_PAGEBOX, wxID_PAGEHIST, wxID_PAGEPROB,
wxID_PAGESUMMARY, wxID_PAGETIMESERIES, wxID_TABPLOTS
] = [wx.NewId() for _init_ctrls in range(7)]
class pnlPlot(fnb.FlatNotebook):
def __init__(self, parent, taskserver):
self.taskserver = taskserver
self._init_ctrls(parent)
self.initPubSub()
self.parent = parent
def _init_ctrls(self, parent):
fnb.FlatNotebook.__init__(self, id=wxID_TABPLOTS, name=u'tabPlots',
parent=parent, pos=wx.Point(0, 0), size=wx.Size(491, 288),
agwStyle=fnb.FNB_NODRAG | fnb.FNB_HIDE_TABS)
# style |= fnb.FNB_HIDE_TABS
# self.book.SetAGWWindowStyleFlag(style)
self.pltTS = plotTimeSeries.plotTimeSeries(id=wxID_PAGETIMESERIES, name='pltTS',
parent=self, pos=wx.Point(0, 0), size=wx.Size(605, 458),
style=wx.TAB_TRAVERSAL)
self.AddPage(self.pltTS, 'TimeSeries')
self.pltProb = plotProbability.plotProb(id=wxID_PAGEPROB, name='pltProb',
parent=self, pos=wx.Point(0, 0), size=wx.Size(605, 458),
style=wx.TAB_TRAVERSAL)
self.AddPage(self.pltProb, 'Probablity')
self.pltHist = plotHistogram.plotHist(id=wxID_PAGEHIST, name='pltHist',
parent=self, pos=wx.Point(0, 0), size=wx.Size(605, 458),
style=wx.TAB_TRAVERSAL)
self.AddPage(self.pltHist, 'Histogram')
self.pltBox = plotBoxWhisker.PlotBox(id=wxID_PAGEBOX, name='pltBox',
parent=self, pos=wx.Point(0, 0), size=wx.Size(605, 458),
style=wx.TAB_TRAVERSAL)
self.AddPage(self.pltBox, 'Box/Whisker')
self.pltSum = plotSummary.plotSummary(id=wxID_PAGESUMMARY, name=u'pltSum',
parent=self, pos=wx.Point(784, 256), size=wx.Size(437, 477),
style=wx.TAB_TRAVERSAL)
self.AddPage(self.pltSum, 'Summary')
self._seriesPlotInfo = None
self.editID = None
self.legendVisible = False
def initPubSub(self):
Publisher.subscribe(self.onDateChanged, "onDateChanged")
Publisher.subscribe(self.onDateFull, "onDateFull")
Publisher.subscribe(self.onPlotType, "onPlotType")
Publisher.subscribe(self.onShowLegend, "onShowLegend")
Publisher.subscribe(self.onNumBins, "onNumBins")
Publisher.subscribe(self.onRemovePlot, "removePlot")
Publisher.subscribe(self.onRemovePlots, "removeMultPlot")
Publisher.subscribe(self.onChangeSelection, "changePlotSelection")
Publisher.subscribe(self.onUpdateValues, "updateValues")
Publisher.subscribe(self.clear, "clearPlot")
def onUpdateValues(self, event):
self.pltTS.updateValues()
def onChangeSelection(self, datetime_list):
self.pltTS.changePlotSelection( datetime_list)
def onNumBins(self, numBins):
self.pltHist.changeNumOfBins(numBins)
def onDateChanged(self, startDate, endDate):
self._seriesPlotInfo.updateDateRange(startDate, endDate)
self.redrawPlots()
def onDateFull(self):
self._seriesPlotInfo.updateDateRange()
self.redrawPlots()
# Reset the date to the full date
def onPlotType(self, event, ptype):
self.pltTS.onPlotType(ptype)
self.pltProb.onPlotType(ptype)
def onShowLegend(self, event, isVisible):
try:
self.pltTS.onShowLegend(isVisible)
self.pltProb.onShowLegend(isVisible)
self.legendVisible = isVisible
except AttributeError:
pass
def stopEdit(self):
self._seriesPlotInfo.stopEditSeries()
self.editID = None
self.pltTS.stopEdit()
self.redrawPlots()
def addEditPlot(self, memDB, seriesID, record_service):
self.record_service = record_service
if not self._seriesPlotInfo:
self._seriesPlotInfo = SeriesPlotInfo(memDB, self.taskserver)
self.editID = seriesID
self._seriesPlotInfo.setEditSeries(self.editID)
self.pltTS.setEdit(self.editID)
self.redrawPlots()
def addPlot(self, memDB, seriesID):
"""
Creates the plot
"""
logger.debug("Adding plot")
Publisher.sendMessage("EnablePlotButton", plot=self.getActivePlotID(), isActive=True)
if not self._seriesPlotInfo:
self._seriesPlotInfo = SeriesPlotInfo(memDB, self.taskserver)
self._seriesPlotInfo.update(seriesID, True)
logger.debug("Redrawing plots")
self.redrawPlots()
def onRemovePlot(self, seriesID):
self._seriesPlotInfo.update(seriesID, False)
self.redrawPlots()
def onRemovePlots(self, seriesIDs):
for series in seriesIDs:
self._seriesPlotInfo.update(series.id, False)
self.redrawPlots()
def redrawPlots(self):
logger.debug("Plot Summary")
self.pltSum.Plot(self._seriesPlotInfo)
logger.debug("Plot Probability")
self.pltProb.Plot(self._seriesPlotInfo)
logger.debug("Plot Boxwhisker")
self.pltBox.Plot(self._seriesPlotInfo)
logger.debug("Plot Timeseries")
self.pltTS.Plot(self._seriesPlotInfo)
logger.debug("Plot Histogram")
self.pltHist.Plot(self._seriesPlotInfo)
self.onShowLegend(event=None, isVisible=self.legendVisible)
maxStart, maxEnd, currStart, currEnd = self._seriesPlotInfo.getDates()
Publisher.sendMessage("resetdate", startDate=maxStart, endDate=maxEnd, currStart=currStart, currEnd=currEnd)
def selectPlot(self, value):
self.SetSelection(value)
def getActivePlotID(self):
return self.GetSelection()
def close(self):
self.pltTS.close()
def clear(self):
"""
:return:
"""
if self._seriesPlotInfo:
for seriesID in self._seriesPlotInfo.getSeriesIDs():
self._seriesPlotInfo.update(seriesID, False)
self.redrawPlots()
| bsd-3-clause |
karstenw/nodebox-pyobjc | examples/Extended Application/sklearn/examples/ensemble/plot_random_forest_regression_multioutput.py | 1 | 3492 | """
============================================================
Comparing random forests and the multi-output meta estimator
============================================================
An example to compare multi-output regression with random forest and
the :ref:`multioutput.MultiOutputRegressor <multiclass>` meta-estimator.
This example illustrates the use of the
:ref:`multioutput.MultiOutputRegressor <multiclass>` meta-estimator
to perform multi-output regression. A random forest regressor is used,
which supports multi-output regression natively, so the results can be
compared.
The random forest regressor will only ever predict values within the
range of observations or closer to zero for each of the targets. As a
result the predictions are biased towards the centre of the circle.
Using a single underlying feature the model learns both the
x and y coordinate as output.
"""
print(__doc__)
# Author: Tim Head <betatim@gmail.com>
#
# License: BSD 3 clause
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from sklearn.multioutput import MultiOutputRegressor
# nodebox section
if __name__ == '__builtin__':
# were in nodebox
import os
import tempfile
W = 800
inset = 20
size(W, 600)
plt.cla()
plt.clf()
plt.close('all')
def tempimage():
fob = tempfile.NamedTemporaryFile(mode='w+b', suffix='.png', delete=False)
fname = fob.name
fob.close()
return fname
imgx = 20
imgy = 0
def pltshow(plt, dpi=150):
global imgx, imgy
temppath = tempimage()
plt.savefig(temppath, dpi=dpi)
dx,dy = imagesize(temppath)
w = min(W,dx)
image(temppath,imgx,imgy,width=w)
imgy = imgy + dy + 20
os.remove(temppath)
size(W, HEIGHT+dy+40)
else:
def pltshow(mplpyplot):
mplpyplot.show()
# nodebox section end
# Create a random dataset
rng = np.random.RandomState(1)
X = np.sort(200 * rng.rand(600, 1) - 100, axis=0)
y = np.array([np.pi * np.sin(X).ravel(), np.pi * np.cos(X).ravel()]).T
y += (0.5 - rng.rand(*y.shape))
X_train, X_test, y_train, y_test = train_test_split(X, y,
train_size=400,
random_state=4)
max_depth = 30
regr_multirf = MultiOutputRegressor(RandomForestRegressor(max_depth=max_depth,
random_state=0))
regr_multirf.fit(X_train, y_train)
regr_rf = RandomForestRegressor(max_depth=max_depth, random_state=2)
regr_rf.fit(X_train, y_train)
# Predict on new data
y_multirf = regr_multirf.predict(X_test)
y_rf = regr_rf.predict(X_test)
# Plot the results
plt.figure()
s = 50
a = 0.4
plt.scatter(y_test[:, 0], y_test[:, 1], edgecolor='k',
c="navy", s=s, marker="s", alpha=a, label="Data")
plt.scatter(y_multirf[:, 0], y_multirf[:, 1], edgecolor='k',
c="cornflowerblue", s=s, alpha=a,
label="Multi RF score=%.2f" % regr_multirf.score(X_test, y_test))
plt.scatter(y_rf[:, 0], y_rf[:, 1], edgecolor='k',
c="c", s=s, marker="^", alpha=a,
label="RF score=%.2f" % regr_rf.score(X_test, y_test))
plt.xlim([-6, 6])
plt.ylim([-6, 6])
plt.xlabel("target 1")
plt.ylabel("target 2")
plt.title("Comparing random forests and the multi-output meta estimator")
plt.legend()
# plt.show()
pltshow(plt)
| mit |
cbertinato/pandas | pandas/tests/test_downstream.py | 1 | 4179 | """
Testing that we work in the downstream packages
"""
import importlib
import subprocess
import sys
import numpy as np # noqa
import pytest
from pandas.compat import PY36
from pandas import DataFrame
from pandas.util import testing as tm
def import_module(name):
# we *only* want to skip if the module is truly not available
# and NOT just an actual import error because of pandas changes
if PY36:
try:
return importlib.import_module(name)
except ModuleNotFoundError: # noqa
pytest.skip("skipping as {} not available".format(name))
else:
try:
return importlib.import_module(name)
except ImportError as e:
if "No module named" in str(e) and name in str(e):
pytest.skip("skipping as {} not available".format(name))
raise
@pytest.fixture
def df():
return DataFrame({'A': [1, 2, 3]})
def test_dask(df):
toolz = import_module('toolz') # noqa
dask = import_module('dask') # noqa
import dask.dataframe as dd
ddf = dd.from_pandas(df, npartitions=3)
assert ddf.A is not None
assert ddf.compute() is not None
def test_xarray(df):
xarray = import_module('xarray') # noqa
assert df.to_xarray() is not None
def test_oo_optimizable():
# GH 21071
subprocess.check_call([sys.executable, "-OO", "-c", "import pandas"])
@tm.network
# Cython import warning
@pytest.mark.filterwarnings("ignore:can't:ImportWarning")
def test_statsmodels():
statsmodels = import_module('statsmodels') # noqa
import statsmodels.api as sm
import statsmodels.formula.api as smf
df = sm.datasets.get_rdataset("Guerry", "HistData").data
smf.ols('Lottery ~ Literacy + np.log(Pop1831)', data=df).fit()
# Cython import warning
@pytest.mark.filterwarnings("ignore:can't:ImportWarning")
def test_scikit_learn(df):
sklearn = import_module('sklearn') # noqa
from sklearn import svm, datasets
digits = datasets.load_digits()
clf = svm.SVC(gamma=0.001, C=100.)
clf.fit(digits.data[:-1], digits.target[:-1])
clf.predict(digits.data[-1:])
# Cython import warning and traitlets
@tm.network
@pytest.mark.filterwarnings("ignore")
def test_seaborn():
seaborn = import_module('seaborn')
tips = seaborn.load_dataset("tips")
seaborn.stripplot(x="day", y="total_bill", data=tips)
def test_pandas_gbq(df):
pandas_gbq = import_module('pandas_gbq') # noqa
@pytest.mark.xfail(reason="0.7.0 pending")
@tm.network
def test_pandas_datareader():
pandas_datareader = import_module('pandas_datareader') # noqa
pandas_datareader.DataReader(
'F', 'quandl', '2017-01-01', '2017-02-01')
# importing from pandas, Cython import warning
@pytest.mark.filterwarnings("ignore:The 'warn':DeprecationWarning")
@pytest.mark.filterwarnings("ignore:pandas.util:DeprecationWarning")
@pytest.mark.filterwarnings("ignore:can't resolve:ImportWarning")
@pytest.mark.skip(reason="gh-25778: geopandas stack issue")
def test_geopandas():
geopandas = import_module('geopandas') # noqa
fp = geopandas.datasets.get_path('naturalearth_lowres')
assert geopandas.read_file(fp) is not None
# Cython import warning
@pytest.mark.filterwarnings("ignore:can't resolve:ImportWarning")
def test_pyarrow(df):
pyarrow = import_module('pyarrow') # noqa
table = pyarrow.Table.from_pandas(df)
result = table.to_pandas()
tm.assert_frame_equal(result, df)
@pytest.mark.xfail(reason="pandas-wheels-50", strict=False)
def test_missing_required_dependency():
# GH 23868
# To ensure proper isolation, we pass these flags
# -S : disable site-packages
# -s : disable user site-packages
# -E : disable PYTHON* env vars, especially PYTHONPATH
# And, that's apparently not enough, so we give up.
# https://github.com/MacPython/pandas-wheels/pull/50
call = ['python', '-sSE', '-c', 'import pandas']
with pytest.raises(subprocess.CalledProcessError) as exc:
subprocess.check_output(call, stderr=subprocess.STDOUT)
output = exc.value.stdout.decode()
for name in ['numpy', 'pytz', 'dateutil']:
assert name in output
| bsd-3-clause |
Reagankm/KnockKnock | venv/lib/python3.4/site-packages/matplotlib/testing/image_util.py | 11 | 3765 | # This module contains some functionality from the Python Imaging
# Library, that has been ported to use Numpy arrays rather than PIL
# Image objects.
# The Python Imaging Library is
# Copyright (c) 1997-2009 by Secret Labs AB
# Copyright (c) 1995-2009 by Fredrik Lundh
# By obtaining, using, and/or copying this software and/or its
# associated documentation, you agree that you have read, understood,
# and will comply with the following terms and conditions:
# Permission to use, copy, modify, and distribute this software and its
# associated documentation for any purpose and without fee is hereby
# granted, provided that the above copyright notice appears in all
# copies, and that both that copyright notice and this permission notice
# appear in supporting documentation, and that the name of Secret Labs
# AB or the author not be used in advertising or publicity pertaining to
# distribution of the software without specific, written prior
# permission.
# SECRET LABS AB AND THE AUTHOR DISCLAIMS ALL WARRANTIES WITH REGARD TO
# THIS SOFTWARE, INCLUDING ALL IMPLIED WARRANTIES OF MERCHANTABILITY AND
# FITNESS. IN NO EVENT SHALL SECRET LABS AB OR THE AUTHOR BE LIABLE FOR
# ANY SPECIAL, INDIRECT OR CONSEQUENTIAL DAMAGES OR ANY DAMAGES
# WHATSOEVER RESULTING FROM LOSS OF USE, DATA OR PROFITS, WHETHER IN AN
# ACTION OF CONTRACT, NEGLIGENCE OR OTHER TORTIOUS ACTION, ARISING OUT
# OF OR IN CONNECTION WITH THE USE OR PERFORMANCE OF THIS SOFTWARE.
from __future__ import (absolute_import, division, print_function,
unicode_literals)
import six
from six.moves import xrange
import numpy as np
from matplotlib.cbook import deprecated, warn_deprecated
warn_deprecated('1.4.0', name='matplotlib.testing.image_util',
obj_type='module')
@deprecated('1.4.0')
def autocontrast(image, cutoff=0):
"""
Maximize image contrast, based on histogram. This completely
ignores the alpha channel.
"""
assert image.dtype == np.uint8
output_image = np.empty((image.shape[0], image.shape[1], 3), np.uint8)
for i in xrange(0, 3):
plane = image[:,:,i]
output_plane = output_image[:,:,i]
h = np.histogram(plane, bins=256)[0]
if cutoff:
# cut off pixels from both ends of the histogram
# get number of pixels
n = 0
for ix in xrange(256):
n = n + h[ix]
# remove cutoff% pixels from the low end
cut = n * cutoff / 100
for lo in range(256):
if cut > h[lo]:
cut = cut - h[lo]
h[lo] = 0
else:
h[lo] = h[lo] - cut
cut = 0
if cut <= 0:
break
# remove cutoff% samples from the hi end
cut = n * cutoff / 100
for hi in xrange(255, -1, -1):
if cut > h[hi]:
cut = cut - h[hi]
h[hi] = 0
else:
h[hi] = h[hi] - cut
cut = 0
if cut <= 0:
break
# find lowest/highest samples after preprocessing
for lo in xrange(256):
if h[lo]:
break
for hi in xrange(255, -1, -1):
if h[hi]:
break
if hi <= lo:
output_plane[:,:] = plane
else:
scale = 255.0 / (hi - lo)
offset = -lo * scale
lut = np.arange(256, dtype=np.float)
lut *= scale
lut += offset
lut = lut.clip(0, 255)
lut = lut.astype(np.uint8)
output_plane[:,:] = lut[plane]
return output_image
| gpl-2.0 |
wateraccounting/wa | Collect/MOD9/DataAccess.py | 1 | 12824 | # -*- coding: utf-8 -*-
"""
Authors: Tim Hessels
UNESCO-IHE 2016
Contact: t.hessels@unesco-ihe.org
Repository: https://github.com/wateraccounting/wa
Module: Collect/MOD9
"""
# import general python modules
import os
import numpy as np
import pandas as pd
import gdal
import urllib
import urllib2
from bs4 import BeautifulSoup
import re
import urlparse
import glob
import requests
from joblib import Parallel, delayed
# Water Accounting modules
import wa
import wa.General.raster_conversions as RC
import wa.General.data_conversions as DC
from wa import WebAccounts
def DownloadData(Dir, Startdate, Enddate, latlim, lonlim, Waitbar, cores, hdf_library, remove_hdf):
"""
This function downloads MOD9 daily data
Keyword arguments:
Dir -- 'C:/file/to/path/'
Startdate -- 'yyyy-mm-dd'
Enddate -- 'yyyy-mm-dd'
latlim -- [ymin, ymax] (values must be between -90 and 90)
lonlim -- [xmin, xmax] (values must be between -180 and 180)
cores -- The number of cores used to run the routine. It can be 'False'
to avoid using parallel computing routines.
Waitbar -- 1 (Default) will print a waitbar
"""
# Check start and end date and otherwise set the date to max
if not Startdate:
Startdate = pd.Timestamp('2000-02-24')
if not Enddate:
Enddate = pd.Timestamp('Now')
# Make an array of the days of which the NDVI is taken
Dates = pd.date_range(Startdate, Enddate, freq = 'D')
# Create Waitbar
if Waitbar == 1:
import wa.Functions.Start.WaitbarConsole as WaitbarConsole
total_amount = len(Dates)
amount = 0
WaitbarConsole.printWaitBar(amount, total_amount, prefix = 'Progress:', suffix = 'Complete', length = 50)
# Check the latitude and longitude and otherwise set lat or lon on greatest extent
if latlim[0] < -90 or latlim[1] > 90:
print 'Latitude above 90N or below 90S is not possible. Value set to maximum'
latlim[0] = np.max(latlim[0], -90)
latlim[1] = np.min(latlim[1], 90)
if lonlim[0] < -180 or lonlim[1] > 180:
print 'Longitude must be between 180E and 180W. Now value is set to maximum'
lonlim[0] = np.max(lonlim[0], -180)
lonlim[1] = np.min(lonlim[1], 180)
# Make directory for the MODIS NDVI data
Dir = Dir.replace("/", os.sep)
output_folder = os.path.join(Dir, 'Reflectance', 'MOD9')
if not os.path.exists(output_folder):
os.makedirs(output_folder)
TilesVertical, TilesHorizontal = wa.Collect.MOD15.DataAccess.Get_tiles_from_txt(output_folder, hdf_library, latlim, lonlim)
# Pass variables to parallel function and run
args = [output_folder, TilesVertical, TilesHorizontal, lonlim, latlim, hdf_library]
if not cores:
for Date in Dates:
RetrieveData(Date, args)
if Waitbar == 1:
amount += 1
WaitbarConsole.printWaitBar(amount, total_amount, prefix = 'Progress:', suffix = 'Complete', length = 50)
results = True
else:
results = Parallel(n_jobs=cores)(delayed(RetrieveData)(Date, args)
for Date in Dates)
if remove_hdf == 1:
# Remove all .hdf files
os.chdir(output_folder)
files = glob.glob("*.hdf")
for f in files:
os.remove(os.path.join(output_folder, f))
# Remove all .txt files
files = glob.glob("*.txt")
for f in files:
os.remove(os.path.join(output_folder, f))
return results
def RetrieveData(Date, args):
"""
This function retrieves MOD9 Reflectance data for a given date from the
http://e4ftl01.cr.usgs.gov/ server.
Keyword arguments:
Date -- 'yyyy-mm-dd'
args -- A list of parameters defined in the DownloadData function.
"""
# Argument
[output_folder, TilesVertical, TilesHorizontal, lonlim, latlim, hdf_library] = args
# Collect the data from the MODIS webpage and returns the data and lat and long in meters of those tiles
try:
Collect_data(TilesHorizontal, TilesVertical, Date, output_folder, hdf_library)
except:
print "Was not able to download the file"
# Define the output name of the collect data function
name_collect = os.path.join(output_folder, 'Merged.tif')
# Reproject the MODIS product to epsg_to
epsg_to ='4326'
name_reprojected = RC.reproject_MODIS(name_collect, epsg_to)
# Clip the data to the users extend
data, geo = RC.clip_data(name_reprojected, latlim, lonlim)
# Save results as Gtiff
ReffileName = os.path.join(output_folder, 'Reflectance_MOD09GQ_-_daily_' + Date.strftime('%Y') + '.' + Date.strftime('%m') + '.' + Date.strftime('%d') + '.tif')
DC.Save_as_tiff(name=ReffileName, data=data, geo=geo, projection='WGS84')
# remove the side products
os.remove(os.path.join(output_folder, name_collect))
os.remove(os.path.join(output_folder, name_reprojected))
return True
def Collect_data(TilesHorizontal,TilesVertical,Date,output_folder, hdf_library):
'''
This function downloads all the needed MODIS tiles from http://e4ftl01.cr.usgs.gov/MOLT/MOD13Q1.006/ as a hdf file.
Keywords arguments:
TilesHorizontal -- [TileMin,TileMax] max and min horizontal tile number
TilesVertical -- [TileMin,TileMax] max and min vertical tile number
Date -- 'yyyy-mm-dd'
output_folder -- 'C:/file/to/path/'
'''
# Make a new tile for the data
sizeX = int((TilesHorizontal[1] - TilesHorizontal[0] + 1) * 4800)
sizeY = int((TilesVertical[1] - TilesVertical[0] + 1) * 4800)
DataTot = np.zeros((sizeY, sizeX))
# Load accounts
username, password = WebAccounts.Accounts(Type = 'NASA')
# Create the Lat and Long of the MODIS tile in meters
for Vertical in range(int(TilesVertical[0]), int(TilesVertical[1])+1):
Distance = 231.65635826395834 # resolution of a MODIS pixel in meter
countY=(TilesVertical[1] - TilesVertical[0] + 1) - (Vertical - TilesVertical[0])
for Horizontal in range(int(TilesHorizontal[0]), int(TilesHorizontal[1]) + 1):
countX=Horizontal - TilesHorizontal[0] + 1
# Download the MODIS NDVI data
url = 'https://e4ftl01.cr.usgs.gov/MOLT/MOD09GQ.006/' + Date.strftime('%Y') + '.' + Date.strftime('%m') + '.' + Date.strftime('%d') + '/'
# Reset the begin parameters for downloading
downloaded = 0
N=0
# Check the library given by user
if hdf_library is not None:
os.chdir(hdf_library)
hdf_name = glob.glob("MOD09GQ.A%s%03s.h%02dv%02d.*" %(Date.strftime('%Y'), Date.strftime('%j'), Horizontal, Vertical))
if len(hdf_name) == 1:
hdf_file = os.path.join(hdf_library, hdf_name[0])
if os.path.exists(hdf_file):
downloaded = 1
file_name = hdf_file
if not downloaded == 1:
# Get files on FTP server
f = urllib2.urlopen(url)
# Sum all the files on the server
soup = BeautifulSoup(f, "lxml")
for i in soup.findAll('a', attrs = {'href': re.compile('(?i)(hdf)$')}):
# Find the file with the wanted tile number
Vfile=str(i)[30:32]
Hfile=str(i)[27:29]
if int(Vfile) is int(Vertical) and int(Hfile) is int(Horizontal):
# Define the whole url name
full_url = urlparse.urljoin(url, i['href'])
# if not downloaded try to download file
while downloaded == 0:
try:# open http and download whole .hdf
nameDownload = full_url
file_name = os.path.join(output_folder,nameDownload.split('/')[-1])
if os.path.isfile(file_name):
downloaded = 1
else:
x = requests.get(nameDownload, allow_redirects = False)
try:
y = requests.get(x.headers['location'], auth = (username, password))
except:
from requests.packages.urllib3.exceptions import InsecureRequestWarning
requests.packages.urllib3.disable_warnings(InsecureRequestWarning)
y = requests.get(x.headers['location'], auth = (username, password), verify = False)
z = open(file_name, 'wb')
z.write(y.content)
z.close()
statinfo = os.stat(file_name)
# Say that download was succesfull
if int(statinfo.st_size) > 10000:
downloaded = 1
# If download was not succesfull
except:
# Try another time
N = N + 1
# Stop trying after 10 times
if N == 10:
print 'Data from ' + Date.strftime('%Y-%m-%d') + ' is not available'
downloaded = 1
try:
# Open .hdf only band with NDVI and collect all tiles to one array
dataset = gdal.Open(file_name)
sdsdict = dataset.GetMetadata('SUBDATASETS')
sdslist = [sdsdict[k] for k in sdsdict.keys() if '_2_NAME' in k]
sds = []
for n in sdslist:
sds.append(gdal.Open(n))
full_layer = [i for i in sdslist if 'sur_refl_b01_1' in i]
idx = sdslist.index(full_layer[0])
if Horizontal == TilesHorizontal[0] and Vertical == TilesVertical[0]:
geo_t = sds[idx].GetGeoTransform()
# get the projection value
proj = sds[idx].GetProjection()
data = sds[idx].ReadAsArray()
countYdata = (TilesVertical[1] - TilesVertical[0] + 2) - countY
DataTot[int((countYdata - 1) * 4800):int(countYdata * 4800), int((countX - 1) * 4800):int(countX * 4800)]=data
del data
# if the tile not exists or cannot be opened, create a nan array with the right projection
except:
if Horizontal==TilesHorizontal[0] and Vertical==TilesVertical[0]:
x1 = (TilesHorizontal[0] - 19) * 4800 * Distance
x4 = (TilesVertical[0] - 9) * 4800 * -1 * Distance
geo = [x1, Distance, 0.0, x4, 0.0, -Distance]
geo_t=tuple(geo)
proj='PROJCS["unnamed",GEOGCS["Unknown datum based upon the custom spheroid",DATUM["Not specified (based on custom spheroid)",SPHEROID["Custom spheroid",6371007.181,0]],PRIMEM["Greenwich",0],UNIT["degree",0.0174532925199433]],PROJECTION["Sinusoidal"],PARAMETER["longitude_of_center",0],PARAMETER["false_easting",0],PARAMETER["false_northing",0],UNIT["Meter",1]]'
data=np.ones((4800,4800)) * (-9999)
countYdata=(TilesVertical[1] - TilesVertical[0] + 2) - countY
DataTot[(countYdata - 1) * 4800:countYdata * 4800,(countX - 1) * 4800:countX * 4800] = data
# Make geotiff file
name2 = os.path.join(output_folder, 'Merged.tif')
driver = gdal.GetDriverByName("GTiff")
dst_ds = driver.Create(name2, DataTot.shape[1], DataTot.shape[0], 1, gdal.GDT_Float32, ['COMPRESS=LZW'])
try:
dst_ds.SetProjection(proj)
except:
proj='PROJCS["unnamed",GEOGCS["Unknown datum based upon the custom spheroid",DATUM["Not specified (based on custom spheroid)",SPHEROID["Custom spheroid",6371007.181,0]],PRIMEM["Greenwich",0],UNIT["degree",0.0174532925199433]],PROJECTION["Sinusoidal"],PARAMETER["longitude_of_center",0],PARAMETER["false_easting",0],PARAMETER["false_northing",0],UNIT["Meter",1]]'
x1 = (TilesHorizontal[0] - 18) * 4800 * Distance
x4 = (TilesVertical[0] - 9) * 4800 * -1 * Distance
geo = [x1, Distance, 0.0, x4, 0.0, -Distance]
geo_t = tuple(geo)
dst_ds.SetProjection(proj)
dst_ds.GetRasterBand(1).SetNoDataValue(-9999)
dst_ds.SetGeoTransform(geo_t)
dst_ds.GetRasterBand(1).WriteArray(DataTot*0.0001)
dst_ds = None
sds = None
return()
| apache-2.0 |
Lab603/PicEncyclopedias | jni-build/jni-build/jni/include/tensorflow/contrib/factorization/python/ops/gmm_test.py | 4 | 6387 | # Copyright 2016 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Tests for ops.gmm."""
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import numpy as np
from six.moves import xrange # pylint: disable=redefined-builtin
import tensorflow as tf
from tensorflow.contrib.factorization.python.ops.gmm import GMM
from tensorflow.contrib.factorization.python.ops.kmeans import KMeansClustering as KMeans
from tensorflow.contrib.learn.python.learn.estimators import run_config
FLAGS = tf.app.flags.FLAGS
class GMMTest(tf.test.TestCase):
def setUp(self):
np.random.seed(3)
tf.set_random_seed(2)
self.num_centers = 2
self.num_dims = 2
self.num_points = 4000
self.batch_size = 100
self.true_centers = self.make_random_centers(self.num_centers,
self.num_dims)
self.points, self.assignments, self.scores = self.make_random_points(
self.true_centers,
self.num_points)
self.true_score = np.add.reduce(self.scores)
# Use initial means from kmeans (just like scikit-learn does).
clusterer = KMeans(num_clusters=self.num_centers)
clusterer.fit(self.points, steps=30)
self.initial_means = clusterer.clusters()
@staticmethod
def make_random_centers(num_centers, num_dims):
return np.round(np.random.rand(num_centers,
num_dims).astype(np.float32) * 500)
@staticmethod
def make_random_points(centers, num_points):
num_centers, num_dims = centers.shape
assignments = np.random.choice(num_centers, num_points)
offsets = np.round(np.random.randn(num_points,
num_dims).astype(np.float32) * 20)
points = centers[assignments] + offsets
means = [np.mean(points[assignments == center], axis=0)
for center in xrange(num_centers)]
covs = [np.cov(points[assignments == center].T)
for center in xrange(num_centers)]
scores = []
for r in xrange(num_points):
scores.append(np.sqrt(np.dot(
np.dot(points[r, :] - means[assignments[r]],
np.linalg.inv(covs[assignments[r]])),
points[r, :] - means[assignments[r]])))
return (points, assignments, scores)
def test_clusters(self):
"""Tests the shape of the clusters."""
gmm = GMM(self.num_centers,
initial_clusters=self.initial_means,
batch_size=self.batch_size,
steps=40,
continue_training=True,
random_seed=4,
config=run_config.RunConfig(tf_random_seed=2))
gmm.fit(x=self.points, steps=0)
clusters = gmm.clusters()
self.assertAllEqual(list(clusters.shape),
[self.num_centers, self.num_dims])
def test_fit(self):
gmm = GMM(self.num_centers,
initial_clusters='random',
batch_size=self.batch_size,
random_seed=4,
config=run_config.RunConfig(tf_random_seed=2))
gmm.fit(x=self.points, steps=1)
score1 = gmm.score(x=self.points)
gmm = GMM(self.num_centers,
initial_clusters='random',
batch_size=self.batch_size,
random_seed=4,
config=run_config.RunConfig(tf_random_seed=2))
gmm.fit(x=self.points, steps=10)
score2 = gmm.score(x=self.points)
self.assertGreater(score1, score2)
self.assertNear(self.true_score, score2, self.true_score * 0.15)
def test_infer(self):
gmm = GMM(self.num_centers,
initial_clusters=self.initial_means,
batch_size=self.batch_size,
steps=40,
continue_training=True,
random_seed=4,
config=run_config.RunConfig(tf_random_seed=2))
gmm.fit(x=self.points, steps=60)
clusters = gmm.clusters()
# Make a small test set
points, true_assignments, true_offsets = (
self.make_random_points(clusters, 40))
assignments = np.ravel(gmm.predict(points))
self.assertAllEqual(true_assignments, assignments)
# Test score
score = gmm.score(points)
self.assertNear(score, np.sum(true_offsets), 4.05)
def _compare_with_sklearn(self, cov_type):
# sklearn version.
iterations = 40
np.random.seed(5)
sklearn_assignments = np.asarray([0, 0, 1, 0, 0, 0, 1, 0, 0, 1])
sklearn_means = np.asarray([[144.83417719, 254.20130341],
[274.38754816, 353.16074346]])
sklearn_covs = np.asarray([[[395.0081194, -4.50389512],
[-4.50389512, 408.27543989]],
[[385.17484203, -31.27834935],
[-31.27834935, 391.74249925]]])
# skflow version.
gmm = GMM(self.num_centers,
initial_clusters=self.initial_means,
covariance_type=cov_type,
batch_size=self.num_points,
steps=iterations,
continue_training=True,
config=run_config.RunConfig(tf_random_seed=2))
gmm.fit(self.points)
skflow_assignments = gmm.predict(self.points[:10, :]).astype(int)
self.assertAllClose(sklearn_assignments,
np.ravel(skflow_assignments))
self.assertAllClose(sklearn_means, gmm.clusters())
if cov_type == 'full':
self.assertAllClose(sklearn_covs, gmm.covariances(), rtol=0.01)
else:
for d in [0, 1]:
self.assertAllClose(np.diag(sklearn_covs[d]),
gmm.covariances()[d, :], rtol=0.01)
def test_compare_full(self):
self._compare_with_sklearn('full')
def test_compare_diag(self):
self._compare_with_sklearn('diag')
if __name__ == '__main__':
tf.test.main()
| mit |
nblago/utils | src/model/BBFit.py | 1 | 66521 | # -*- coding: utf-8 -*-
"""
Created on Thu Feb 22 10:57:34 2018
Class that enables to fit a black body function to a set of magntidues.
@author: nadiablago
@version: 0.22
"""
from __future__ import print_function
import matplotlib
from matplotlib import pylab as plt
import corner
from astropy import units as u
import astropy.constants as cnt
import os, sys
import numpy as np
import emcee
from scipy import stats
import extinction
from astropy.cosmology import FlatLambdaCDM
import warnings
#If PYSYN_CDBS is not defined, it adds the environment variable which points to the
#filter response files for the bands we are interested in.
if not 'PYSYN_CDBS' in os.environ.keys():
print ("Adding the Pysynphot environment:")
os.environ['PYSYN_CDBS'] = "/Users/USER/SOMEWHERE/pysynphot_files"
print ('PYSYN_CDBS environment variable set to: ', os.environ['PYSYN_CDBS'])
'''os.environ['PYSYN_CDBS'] = "/scratch/Software/pysynphot_files/cdbs/"
# Add the environment variable which points to the filter response files for the bands we are interested in.
if not 'PYSYN_CDBS' in os.environ.keys():
print("Adding the Pysynphot environment:")
os.environ['PYSYN_CDBS'] = "/scratch/Software/pysynphot_files/cdbs/"
print('PYSYN_CDBS environment variable set to: ', os.environ['PYSYN_CDBS'])'''
os.environ['PYSYN_CDBS'] = "/Users/nadiablago/Documents/Software/pysynphot_files/"
import pysynphot as ps
class BBFit:
def __init__(self):
'''
Constructor initializes all the parameters to
defaults.
'''
#Some predefined constants in the units we need them
self.c = cnt.c.to(u.cm/u.s).value #2.99792458e+10 #cm / s
self.h = cnt.h.to(u.erg * u.s).value #6.62607004e-27 #erg s
self.k_B = cnt.k_B.to(u.erg / u.K).value#1.38064852e-16 #erg / K
#Source parameters
self.av_host = 0
self.av_mw = 0
self.law = "Fitzpatrick"
self.law_mw = "Fitzpatrick"
#Black body models
self.initT1 = 10000 #K
self.initR1 = 1 # Rsun
self.initT2 = 3000 #K
self.initR2 = 1 # Rsun
self.z = None
self.distMpc = None #in Mpc
self.mjd = 0
#Power law models
self.alpha = 0.75
self.alphaerr1 = 0
self.alphaerr2 = 0
self.scale = 1
self.scaleerr1 = 0.1
self.scaleerr2 = 0.1
#Disk model (scale is already in the power law model)
#Stellar mass, radius, log accretion mass per year, outer radius of accretion disk
self.Mstar = 1
self.Mstarerr1 = 0.1
self.Mstarerr2 = 0.1
self.Rstar = 1
self.Rstarerr1 = 0.1
self.rstarerr2 = 0.1
self.logMacc = -8
self.logMaccerr1 = -9
self.logMaccerr2 = -9
self.R_out = 3
self.R_outerr1 = 1
self.R_outerr2 = 1
#Location for plots
self.plotdir = "../../data/plots"
#Location for fit results
self.resdir = "../../data/modelfits"
self.resfile = "fit_results.txt"
#MCMC parameters
self.method = 'ensemble' #or HA for Hastings
self.mhtune = True # tuning of the Metropolis-Hastings
self.niterations = 10000
self.burnin = 5000
self.threads = 10
self.nwalkers = 20
self.sampler = None
self.model = "BlackBody" #others are "BlackBody_Av" or "BlackBody2_Av", "PowerLaw", "PowerLaw_BlackBody"
#Input data parameters.
#The fitter will run either with magnitudes or with fluxes
self.mags = None
self.magerrs = None
self.bands = None
#Indicates whether the magnitude is in AB or Vega
self.photsys = None
self.wls = None
self.fluxes = None
self.fluxerrs = None
#Output
self.T = None
self.Terr1 = None
self.Terr2 = None
self.R = None
self.Rerr1 = None
self.Rerr2 = None
self.L = None
self.Lerr1 = None
self.Lerr2 = None
#Output for the secondary star
self.Tsec = None
self.Tsecerr1 = None
self.Tsecerr2 = None
self.Rsec = None
self.Rsecerr1 = None
self.Rsecerr2 = None
self.Lsec = None
self.Lsecerr1 = None
self.Lsecerr2 = None
self.cosmo = FlatLambdaCDM(H0=70, Om0=0.3, Tcmb0=2.725)
#Set the plotting characteristics
self._matplotlib_init()
self.banddic = {"Y": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/ctio_y_andicam.dat"),
"J": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/bessell_j_002.fits"),
"H": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/bessell_h_002.fits"),
"K": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/bessell_k_002.fits"),
"keck,J": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Keck_NIRC2.J.dat"),
"keck,H": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Keck_NIRC2.H.dat"),
"keck,Ks": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Keck_NIRC2.Ks.dat"),
"keck,K": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Keck_NIRC2.K.dat"),
"spitzer,3.6": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Spitzer_irac1_3.6.dat"),
"spitzer,4.5": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Spitzer_irac2_4.5.dat"),
"spitzer,5.8": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Spitzer_irac3_5.8.dat"),
"spitzer,8.0": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Spitzer_irac4_8.0.dat"),
"wise,w1": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/WISE_WISE.W1.dat"),
"wise,w2": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/WISE_WISE.W2.dat"),
"wise,w3": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/WISE_WISE.W3.dat"),
"wise,w4": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/WISE_WISE.W4.dat"),
"swift,uvw2": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/swift_uvw2_uvot.dat"),
"swift,uvm2": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/swift_uvm2_uvot.dat"),
"swift,uvw1": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/swift_uvw1_uvot.dat"),
"swift,u": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/swift_u_uvot.dat"),
"swift,b": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/swift_b_uvot.dat"),
"swift,v": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/swift_v_uvot.dat"),
"paranal,Y": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Paranal_VISTA.Y.dat"),
"paranal,Z": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Paranal_VISTA.Z.dat"),
"paranal,J": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Paranal_VISTA.J.dat"),
"paranal,H": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Paranal_VISTA.H.dat"),
"paranal,Ks": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Paranal_VISTA.Ks.dat"),
"omegacam,u": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Paranal_OmegaCAM.u_SDSS.dat"),
"omegacam,g": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Paranal_OmegaCAM.g_SDSS.dat"),
"omegacam,r": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Paranal_OmegaCAM.r_SDSS.dat"),
"omegacam,i": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Paranal_OmegaCAM.i_SDSS.dat"),
"omegacam,z": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Paranal_OmegaCAM.z_SDSS.dat"),
"omegacam,Halpha": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Paranal_OmegaCAM.Halpha.dat"),
"nirc2,j": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Keck_NIRC2.J.dat"),
"nirc2,h": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Keck_NIRC2.H.dat"),
"nirc2,ks": os.path.join(os.environ['PYSYN_CDBS'], "comp/nonhst/Keck_NIRC2.Ks.dat")
}
def _matplotlib_init(self):
'''
Set up preferences on matplotlib plot appearance.
'''
matplotlib.rcParams['xtick.minor.size'] = 6
matplotlib.rcParams['xtick.major.size'] = 6
matplotlib.rcParams['ytick.major.size'] = 6
matplotlib.rcParams['xtick.minor.size'] = 4
matplotlib.rcParams['ytick.minor.size'] = 4
matplotlib.rcParams['lines.linewidth'] = 0.5
matplotlib.rcParams['axes.linewidth'] = 1.5
matplotlib.rcParams['font.size']= 14.0
matplotlib.rcParams['font.family']= 'sans-serif'
matplotlib.rcParams['xtick.major.width']= 2.
matplotlib.rcParams['ytick.major.width']= 2.
matplotlib.rcParams['ytick.direction']='in'
matplotlib.rcParams['xtick.direction']='in'
def _band2flux(self):
'''
Will transform the magnitude measurement into a flux measurement.
'''
wls = np.array([])
fluxes = np.array([])
fluxerr = np.array([])
#Create a black body spectrum with an arbitrary value
lam = np.linspace(100, 120000, 10000)
sp = ps.BlackBody(10000)
sp.convert('flam')
sp2 = self._model_2(lam, 10000, 1)
sp2 = sp2 * np.max(sp.flux) / np.max(sp2)
sp = ps.ArraySpectrum(lam, sp2)
for b, m, me, psys in zip(self.bands, self.mags, self.magerrs, self.photsys):
print ("Band,",b)
#Create the observation bandpass
try:
band = ps.ObsBandpass(b)
except ValueError:
#The band is not in the standard list
#We need to go to the dictionary to retrieve the transmission function.
band = ps.FileBandpass(self.banddic[b])
#band.waveunits.convert("angstrom")
#else:
# band.waveunits = ps.units.Angstrom
#Oftain the effective (average) wavelength
effwave = band.avgwave()
#Correct for Milky Way extinction
m = m - extinction.fitzpatrick99(np.array([effwave]), a_v=self.av_mw, unit='aa')[0]
#Normalize the spectrum to the magnitude of the observation
sp_norm = sp.renorm(m, psys, band, force="extrap")
#Observe with the band
obs = ps.Observation(sp_norm, band)
#Get the flux
flux = obs.effstim('flam')
wls = np.append(wls, effwave)
fluxes = np.append(fluxes, flux)
#Compute the error bars
flux_high = flux * 10**(0.4*me)
flux_low = flux * 10**(-0.4*me)
fluxerr = np.append(fluxerr, np.average([flux - flux_low, flux_high-flux]))
return wls, fluxes, fluxerr
def _model(self, lam, p):
'''
Returns the flux for the single BlackBody model for the wavelength introduced.
lam is in A.
p = (T, R)
'''
lam = lam * u.Angstrom
T = p[0] * u.K
R = (p[1] * u.Rsun).to(u.cm)
with warnings.catch_warnings():
warnings.simplefilter("ignore")
Area = np.pi * (4 * np.pi * R**2)
flam = Area * (2*cnt.h*((cnt.c).to(u.cm/u.s))**2/( (lam.to(u.cm))**5))/ \
(np.exp((cnt.h*cnt.c)/(lam.to(u.m)*cnt.k_B*T))-1)
return flam.to(u.erg/u.s/u.Angstrom).value
def _model_2(self, lam, T, R):
'''
Return units: erg s-1 A-1
As we multiply by the area of the emitting source (in cm**2)
'''
lam = lam * u.Angstrom
T = T * u.K
R = (R * u.Rsun).to(u.cm)
Area = np.pi * (4 * np.pi * R**2)
flam = Area * (2*cnt.h*((cnt.c).to(u.cm/u.s))**2/( (lam.to(u.cm))**5))/ \
(np.exp((cnt.h*cnt.c)/(lam.to(u.m)*cnt.k_B*T))-1)
return flam.to(u.erg/u.s/u.Angstrom).value
def _model_av_r(self, lam, p):
'''
Return units: erg s-1 A-1
As we multiply by the area of the emitting source (in cm**2)
'''
T = p[0] * u.K
R = (p[1] * u.Rsun).to(u.cm)
a_v = p[2]
if a_v < 0:
return lam * np.inf
#Compute the effect of reddening as a flux factor
flux_red = 10**(-0.4 * extinction.fitzpatrick99(lam, a_v, unit='aa'))
lam = lam * u.Angstrom
area = np.pi * (4 * np.pi * R**2)
flam = area * (2*cnt.h*((cnt.c).to(u.cm/u.s))**2/( (lam.to(u.cm))**5))/ \
(np.exp((cnt.h*cnt.c)/(lam.to(u.m)*cnt.k_B*T))-1)
#Apply the reddening
flam = flam.to(u.erg/u.s/u.Angstrom).value * flux_red
return flam
def _model_av_r_2(self, lam, T, R, a_v):
'''
Return units: erg s-1 A-1
'''
return self._model_av_r(lam, (T, R, a_v))
def _model2_av(self, lam, p):
'''
Return units: erg s-1 A-1
As we multiply by the area of the emitting source (in cm**2)
'''
T1 = p[0] * u.K
R1 = (p[1] * u.Rsun).to(u.cm)
a_v = p[2]
T2 = p[3] * u.K
R2 = (p[4] * u.Rsun).to(u.cm)
#Compute the effect of reddening as a flux factor
flux_red = 10**(-0.4 * extinction.fitzpatrick99(lam, a_v, unit='aa'))
lam = lam * u.Angstrom
area1 = np.pi * (4 * np.pi * R1**2)
area2 = np.pi * (4 * np.pi * R2**2)
with warnings.catch_warnings():
warnings.simplefilter("ignore")
flam1 = area1 * (2*cnt.h*((cnt.c).to(u.cm/u.s))**2/( (lam.to(u.cm))**5))/ \
(np.exp((cnt.h*cnt.c)/(lam.to(u.m)*cnt.k_B*T1))-1)
flam2 = area2 * (2*cnt.h*((cnt.c).to(u.cm/u.s))**2/( (lam.to(u.cm))**5))/ \
(np.exp((cnt.h*cnt.c)/(lam.to(u.m)*cnt.k_B*T2))-1)
flam = flam1 + flam2
#Apply the reddening
flam = flam.to(u.erg/u.s/u.Angstrom).value * flux_red
return flam
def _model2_av_2(self, lam, T1, R1, a_v, T2, R2):
'''
Return units: erg s-1 A-1
As we multiply by the area of the emitting source (in cm**2)
'''
return self._model2_av(lam, (T1, R1, a_v, T2, R2))
def _model2_av_r(self, lam, p):
'''
Return units: erg s-1 A-1
'''
T1 = p[0] #In K
R1 = p[1]*69570000000.0 #From Rsun to cm
a_v = p[2]
T2 = p[3]
R2 = p[4]*69570000000.0 #From Rsun to cm
lam = lam * 1e-8 #To cm
if a_v < 0:
return lam * np.inf
#We need an extra pi as it is integrated across all steradians
#The second factor is the surface of the black body
#The third ones is the Plank law
with warnings.catch_warnings():
warnings.simplefilter("ignore")
flam1 = np.pi * (4 * np.pi * R1**2) * ( (2*self.h*self.c**2)/( lam**5))/ (np.exp((self.h*self.c)/(lam*self.k_B*T1))-1)
flam2 = np.pi * (4 * np.pi * R2**2) * ( (2*self.h*self.c**2)/( lam**5))/ (np.exp((self.h*self.c)/(lam*self.k_B*T2))-1)
#Compute the effect of reddening as a flux factor
flux_red = 10**(-0.4 * extinction.fitzpatrick99(lam*1e8, a_v, unit='aa'))
flam = (flam1 + flam2) * flux_red *1e-8 #to erg / s / A
#Apply the reddening and transform to erg /s/ A from cm
return flam
def _model2_av_r_2(self, lam, T1, R1, a_v, T2, R2):
'''
Return units: erg s-1 A-1
As we multiply by the area of the emitting source (in cm**2)
'''
return self._model2_av_r(lam, (T1, R1, a_v, T2, R2))
def _model2_r(self, lam, p):
'''
Return units: erg s-1 A-1
'''
T1 = p[0] #In K
R1 = p[1]*69570000000.0 #From Rsun to cm
T2 = p[2]
R2 = p[3]*69570000000.0 #From Rsun to cm
lam = lam * 1e-8 #To cm
#We need an extra pi as it is integrated across all steradians
#The second factor is the surface of the black body
#The third ones is the Plank law
with warnings.catch_warnings():
warnings.simplefilter("ignore")
flam1 = np.pi * (4 * np.pi * R1**2) * ( (2*self.h*self.c**2)/( lam**5))/ (np.exp((self.h*self.c)/(lam*self.k_B*T1))-1)
flam2 = np.pi * (4 * np.pi * R2**2) * ( (2*self.h*self.c**2)/( lam**5))/ (np.exp((self.h*self.c)/(lam*self.k_B*T2))-1)
flam = (flam1 + flam2)*1e-8 #to erg / s / A
return flam
def _model2_r_2(self, lam, T1, R1, T2, R2):
'''
Return units: erg s-1 A-1
As we multiply by the area of the emitting source (in cm**2)
'''
return self._model2_r(lam, (T1, R1, T2, R2))
def _model_powerlaw(self, lam, p):
'''
Return units: erg s-1 A-1
As we multiply by the area of the emitting source (in cm**2)
'''
lam = lam * u.Angstrom
w0 = 4000 #p[0] #Refernce wavelength
alpha = p[0]
scale = p[1]
a_v = p[2]
f = ps.PowerLaw(w0, alpha)
f.convert('flam')
flam = np.interp(lam, f.wave, f.flux)
flux_red = 10**(-0.4 * extinction.fitzpatrick99(lam, a_v, unit='aa'))
area = 10**scale
return area * flam * flux_red #.to(u.erg/u.s/u.Angstrom).value
def _model_powerlaw_2(self, lam, alpha, scale, a_v):
'''
Return units: erg s-1 A-1
As we multiply by the area of the emitting source (in cm**2)
'''
return self._model_powerlaw(lam, (alpha, scale, a_v))
def _model_powerlaw_bb(self, lam, p):
'''
Return units: erg s-1 A-1
As we multiply by the area of the emitting source (in cm**2)
'''
w0 = 4000 #p[0] #Refernce wavelength
alpha = p[0]
scale = p[1]
T_bb = p[2]
R_bb = p[3]
bb_flux = self._model_2(lam, T_bb, R_bb)
lam = lam * u.Angstrom
f = ps.PowerLaw(w0, alpha)
f.convert('flam')
flam = np.interp(lam, f.wave, f.flux)
area = 10**scale
return area * flam + bb_flux
def _model_powerlaw_bb_2(self, lam, alpha, scale, T_bb, R_bb):
'''
Return units: erg s-1 A-1
'''
return self._model_powerlaw_bb(lam, (alpha, scale, T_bb, R_bb))
def _model_accretion_disk_old2(self, lam, Mstar, Rstar, logMacc, scale, R_out):
'''
Return units: erg s-1 A-1
As we multiply by the area of the emitting source (in cm**2)
'''
return self._model_accretion_disk_old(lam, (Mstar, Rstar, logMacc, scale, R_out))
def _model_accretion_disk_old(self, lam, p):
'''
Equation 1 from Kenyon, Hartmann, Hewett 1988.
'''
Mstar = p[0]
Rstar = p[1]
Macc = p[2]
scale = p[3]
R_out = p[4]
if Mstar<0 or Macc<-12 or Rstar<0.001 or scale<0 or R_out < Rstar:
return np.ones(len(lam))*np.inf
Macc = 10**Macc
R = np.linspace(Rstar,R_out,20)
dR = R[1] - R[0]
F_r = (3 * cnt.G * Mstar * u.Msun * Macc * u.Msun/u.year / 8 / np.pi / (u.Rsun*Rstar)**3) * (Rstar/R)**3 * (1 - (Rstar/R)**0.5)
F_r = F_r.to(u.erg/u.cm**2/u.s)
T_r = ((F_r / cnt.sigma_sb)**0.25).to(u.K)
T_max = 13000 * u.K *(Mstar)**0.25 * (Macc / 1e-5)**0.25 * (Rstar)**-0.75
#Cretae the disk model
#For each differential radii, we compute the black body spectra corresponding
# to the temperature at that radius, and scale it by the flux expected at that
# radius.
disk_model = []
for i, ri in enumerate(R):
if ri>Rstar and ri<=1.5*Rstar:
sp = ps.BlackBody(T_max.value)
#sp = ps.BlackBody(T_r[i].value)
else:
sp = ps.BlackBody(T_r[i].value)
sp.convert('flam')
tot_flux = sp.trapezoidIntegration(sp.wave, sp.flux)
#Compute the total emitted flux for the spherical area.
#Adopt the outer radius as the
dist_flux_fac = np.pi * ((ri+dR)**2 - ri**2) * (u.Rsun.to(u.cm))**2
scaled_flux = sp.flux / tot_flux * F_r[i].value #* dist_flux_fac
disk_model.append(scaled_flux)
disk = np.array(disk_model)
disk = np.nansum(disk, axis=0)
sp = ps.ArraySpectrum(sp.wave, disk)
#int_flux = sp.trapezoidIntegration(sp.wave, sp.flux)
int_flux = np.max(sp.flux)
#Normalize (recover) the integral flux from 1kpc
flux_norm= sp.flux #/int_flux
#sp_norm = ps.ArraySpectrum(sp.wave, flux_norm)
flux_norm = np.interp(lam, sp.wave, flux_norm)
#flux_red = 10**(-0.4 * extinction.fitzpatrick99(lam, a_v, unit='aa'))
return flux_norm #* scale #* flux_red
def _model_disk_T(self, R, Mstar, Rstar, logMacc):
F_r = (3 * cnt.G * Mstar * 10**float(logMacc) * (u.Msun**2/u.year)) \
/ (8 * np.pi * (u.Rsun*R)**3) \
* (1 - (Rstar/R)**0.5)
T_r = ((F_r / cnt.sigma_sb)**0.25).to(u.K)
#print (F_r, T_r)
mask = (R>=Rstar) * (R<=1.5*Rstar)
if np.count_nonzero(mask)>0:
T_max = 13000 * u.K *(Mstar)**0.25 * (10**float(logMacc) / 1e-5)**0.25 * (Rstar)**-0.75
T_r[mask] = T_max
#print (mask, "Tmax", T_max, np.count_nonzero(mask))
return T_r.value
def _model_accretion_disk2(self, lam, Mstar, Rstar, logMacc, R_out):
'''
Return units: erg s-1 A-1
As we multiply by the area of the emitting source (in cm**2)
'''
return self._model_accretion_disk(lam, (Mstar, Rstar, logMacc, R_out))
def _model_accretion_disk(self, lam, p):
Mstar = np.maximum(1e-6, p[0])
Rstar = np.maximum(1e-6, p[1])
logMacc = np.maximum(-12, np.minimum(-7, p[2]))
R_out = np.maximum(1e-6, p[3])
i = 45.0
#Deg to radians
i = np.deg2rad(i%360)
d = self.distMpc*(u.Mpc).to(u.cm)
R = np.linspace(Rstar, R_out, 30)*u.Rsun
nu = (cnt.c / (lam*u.Angstrom)).to(u.Hz)
T_r = self._model_disk_T(R.value, Mstar, Rstar, logMacc)
F_nu_arr = []
for ni in nu:
I_nu_r = R / (np.exp(cnt.h * ni/(cnt.k_B*T_r*u.K)) - 1)
I_flux = np.trapz(I_nu_r, R)
F_nu = (4 * np.pi * cnt.h * np.cos(i)*ni**3)/(cnt.c**2 * d**2) * I_flux
F_nu_arr.append(F_nu.to(u.erg/u.s/u.Hz).value)
F_nu_arr = np.array(F_nu_arr)
s = ps.ArraySpectrum(lam, F_nu_arr, fluxunits='fnu', waveunits='Angstrom')
s.convert('flam')
fluxFactor = 4*np.pi*d**2
return s.flux*fluxFactor
def _get_Qnu(self, a, lam, wavedusttype="silicate"):
'''
'''
from scipy import interpolate
x = np.array([0.001, 0.01, 0.1, 1]) #size
y = np.array([0.01, 0.06, 0.2, 7, 10 ]) #wavelength
#--> size
# | wave
# v
z = np.array([[0.02, 0.2, 0.85, 0.85],
[0.02, 0.7, 0.7, 0.7],
[0.001, 0.01, 0.7, 0.7],
[0.00007, 0.001, 0.01, 0.1],
[0.001, 0.01, 0.1, 1]])
f = interpolate.interp2d(x, y, z, kind='linear')
return f(a, lam)
def _get_knu(self, a, wave, rho=1, ):
'''
Returns the values for the dust mass absorption coefficient
for the Spitzer bands for the given grain size and wavelength.
k_nu = (3. / 4 * np.pi * rho * a**3)* (np.pi * a**2 * Q_nu(a))
'''
k_nu = (3. / 4 * np.pi * rho * a**3)* (np.pi * a**2 * self.Q_nu(a, wave))
return k_nu
def _model_dust(self, Md, Td, a):
'''
Using the dust modelling approach from Fox et. al. 2010.
The assumption is that the dust is optically thin and that there is only one size and
one dust composition.
The opactities are taken from their Figure 4 values.
F_nu = M_d B_nu (T_d )k_nu(a) / d**2
'''
Bnu = ps.BlackBody(Td)
Bnu.convert('fnu')
knu = self._get_knu(a, wave) * u.cm**2 / u.g
Fnu = Md * u.Msun * Bnu * knu / (self.distMpc * u.Mpc)**2
#likelihood function
def _like(self, p, xdat, ydat, errdat, debug=False):
'''
p: function parameters
args: carry anything we want to pass to our function (e.g. the data)
'''
if self.model == "BlackBody":
ymod = self._model(xdat, p)
elif self.model == "BlackBody_Av":
ymod = self._model_av_r(xdat, p)
elif self.model == "BlackBody2_Av":
ymod = self._model2_av_r(xdat, p)
elif self.model == "BlackBody2":
ymod = self._model2_r(xdat, p)
elif self.model == "PowerLaw":
ymod = self._model_powerlaw(xdat, p)
elif self.model == "PowerLaw_BlackBody":
ymod = self._model_powerlaw_bb(xdat, p)
elif self.model == "Disk":
ymod = self._model_accretion_disk(xdat, p)
else:
print ("Unknown model", self.model)
return np.nan
#Discard models which exceed the upper limits
if (np.any(ymod[errdat<0] > ydat[errdat<0])):
prob = 1e-320
#Compute the likelihood with only valid datapoints.
else:
prob = stats.norm.pdf(ydat[errdat>0] , ymod[errdat>0] , errdat[errdat>0] )
# log probabilities
# we add tiny number to avoid NaNs
mylike = np.log(prob + 1e-320).sum()
return mylike
def _logposterior(self, p, xdat, ydat, errdat):
'''
Returns the posterior of the observations. In essence the likelihood and the prior:
#log(likelihood) + log(prior)
'''
lp = self._logprior(p)
if (not np.isinf(lp)):
lp= self._like(p, xdat, ydat, errdat) + lp
return lp
def _logprior(self, p):
'''
Returns the prior probability distribution for each model.
'''
if self.model == "BlackBody":
T1 = p[0]
R1 = p[1]
if T1 < 0 or R1 < 0:
return -np.inf
logp = stats.uniform.logpdf(T1, 10, 15000)
logp = logp + stats.uniform.logpdf(R1, 1, 50000)
if self.model =="BlackBody_Av":
T1 = p[0]
R1 = p[1]
av = p[2]
if T1 < 0 or R1 < 0 or av < 0:
return -np.inf
else:
logp = stats.uniform.logpdf(T1, 10, 15000)
logp = logp + stats.uniform.logpdf(R1, 10000, 120000)
logp = logp + stats.uniform.logpdf(av, 0, 3)
elif self.model == "BlackBody2":
T1 = p[0]
R1 = p[1]
T2 = p[2]
R2 = p[3]
if T1 < 0 or T2 > T1 or T2 < 0 or R1 < 0 or R2<0:
return - np.inf
else:
logp = stats.uniform.logpdf(T1, 100, 10000)
logp = logp + stats.uniform.logpdf(R1, 10, 12000)
logp = logp + stats.uniform.logpdf(T2, 10, 5000)
logp = logp + stats.uniform.logpdf(R2, 10, 12000)
elif self.model == "BlackBody2_Av":
T1 = p[0]
R1 = p[1]
av = p[2]
T2 = p[3]
R2 = p[4]
if T1 < 0 or T2 > T1 or T2 < 0 or av < 0 or av > 10:
return - np.inf
else:
logp = stats.uniform.logpdf(T1, 100, 1000)
logp = logp + stats.uniform.logpdf(R1, 10000, 120000)
logp = logp + stats.uniform.logpdf(av, 0, 3)
logp = logp + stats.uniform.logpdf(T2, 100, 1000)
logp = logp + stats.uniform.logpdf(R2, 10000, 120000)
elif self.model == "PowerLaw":
alpha = p[0]
scale = p[1]
av = p[2]
if av < 0:
logp = -np.inf
else:
logp = stats.uniform.logpdf(alpha, 0, 3)
logp = logp + stats.uniform.logpdf(scale, 0.1, 100)
logp = logp + stats.uniform.logpdf(av, 0, 3)
elif self.model == "PowerLaw_BlackBody":
alpha = p[0]
scale = p[1]
T1 = p[2]
R1 = p[3]
if R1 < 0 or T1 < 0 or alpha < 0:
logp = -np.inf
else:
logp = stats.uniform.logpdf(alpha, 0, 3)
logp = logp + stats.uniform.logpdf(scale, 0.1, 100)
logp = logp + stats.uniform.logpdf(T1, 500, 20000)
logp = logp + stats.uniform.logpdf(R1, 0, 500)
elif self.model == "Disk":
Mstar = p[0]
Rstar = p[1]
logMacc = p[2]
R_out = p[3]
if Rstar < 0 or Mstar < 0 or logMacc < -12 or R_out<0 or R_out < Rstar:
logp = -np.inf
else:
logp = stats.uniform.logpdf(Mstar, 0, 1.44)
logp = logp + stats.uniform.logpdf(Rstar, 0, 10)
logp = logp + stats.uniform.logpdf(logMacc, -12, 7)
logp = logp + stats.uniform.logpdf(R_out, 0, 50)
return logp
def _get_max_and_intervals(self, x):
'''
Provided a chain of samples, finds the average value and returns the values
for a 1 sigma distribution following the 34 and 66 percentiles.
'''
return np.percentile(x, 34), np.percentile(x, 50), np.percentile(x, 66)
#return percent1, maxp, percent2
def _area2rsun(self, A):
'''
Given the area of the black body in cm2 returns the radius for the object in solar radius.
'''
Aream2 = A * u.cm**2 # add units
Rad = np.sqrt(Aream2/(4*(np.pi)**2)).to(u.Rsun) #in Rsun
return Rad.value
def _fill_output(self):
'''
Computes the confidence intervals from the MCMC distribution.
Transforms the temperature ad radius into a black body luminosity.
'''
if self.model.startswith("BlackBody"):
T1, T, T2 = self._get_max_and_intervals(self.sampler.flatchain[:,0])
R1, R, R2 = self._get_max_and_intervals(self.sampler.flatchain[:,1])
self.T = T
self.Terr1 = T - T1
self.Terr2 = T2 - T
self.R = R
self.Rerr1 = R - R1
self.Rerr2 = R2 - R
self.L = self._get_bol_lum(T, R)
self.Lerr1 = self.L - self._get_bol_lum(T1, R1)
self.Lerr2 = self._get_bol_lum(T2, R2) - self.L
if self.model == "BlackBody_Av":
Av1, Av, Av2 = self._get_max_and_intervals(self.sampler.flatchain[:,2])
self.Av = Av
self.Averr1 = Av - Av1
self.Averr2 = Av2 - Av
elif self.model == "BlackBody2_Av":
Av1, Av, Av2 = self._get_max_and_intervals(self.sampler.flatchain[:,2])
Tsec1, Tsec, Tsec2 = self._get_max_and_intervals(self.sampler.flatchain[:,3])
Rsec1, Rsec, Rsec2 = self._get_max_and_intervals(self.sampler.flatchain[:,4])
self.Av = Av
self.Averr1 = Av - Av1
self.Averr2 = Av2 - Av
self.Tsec = Tsec
self.Tsecerr1 = Tsec - Tsec1
self.Tsecerr2 = Tsec2 - Tsec
self.Rsec = Rsec
self.Rsecerr1 = Rsec - Rsec1
self.Rsecerr2 = Rsec2 - Rsec
elif self.model == "BlackBody2":
Tsec1, Tsec, Tsec2 = self._get_max_and_intervals(self.sampler.flatchain[:,2])
Rsec1, Rsec, Rsec2 = self._get_max_and_intervals(self.sampler.flatchain[:,3])
self.Tsec = Tsec
self.Tsecerr1 = Tsec - Tsec1
self.Tsecerr2 = Tsec2 - Tsec
self.Rsec = Rsec
self.Rsecerr1 = Rsec - Rsec1
self.Rsecerr2 = Rsec2 - Rsec
self.Lsec = self._get_bol_lum(Tsec, Rsec)
self.Lsecerr1 = self.Lsec - self._get_bol_lum(Tsec1, Rsec1)
self.Lsecerr2 = self._get_bol_lum(Tsec2, Rsec2) - self.Lsec
elif self.model=="PowerLaw":
alpha1, alpha, alpha2 = self._get_max_and_intervals(self.sampler.flatchain[:,0])
R1, R, R2 = self._get_max_and_intervals(self.sampler.flatchain[:,1])
Av1, Av, Av2 = self._get_max_and_intervals(self.sampler.flatchain[:,2])
self.alpha = alpha
self.alphaerr1 = alpha - alpha1
self.alphaerr2 = alpha2 - alpha
self.R = R
self.Rerr1 = R - R1
self.Rerr2 = R2 - R
self.Av = Av
self.Averr1 = Av - Av1
self.Averr2 = Av2 - Av
elif self.model=="PowerLaw_BlackBody":
alpha1, alpha, alpha2 = self._get_max_and_intervals(self.sampler.flatchain[:,0])
scale1, scale, scale2 = self._get_max_and_intervals(self.sampler.flatchain[:,1])
T1, T, T2 = self._get_max_and_intervals(self.sampler.flatchain[:,2])
R1, R, R2 = self._get_max_and_intervals(self.sampler.flatchain[:,3])
self.alpha = alpha
self.alphaerr1 = alpha - alpha1
self.alphaerr2 = alpha2 - alpha
self.scale = scale
self.scaleerr1 = scale - scale1
self.scaleerr2 = scale2 - scale
self.T = T
self.Terr1 = T - T1
self.Terr2 = T2 - T
self.R = R
self.Rerr1 = R - R1
self.Rerr2 = R2 - R
elif self.model=="Disk":
Mstar1, Mstar, Mstar2 = self._get_max_and_intervals(self.sampler.flatchain[:,0])
Rstar1, Rstar, Rstar2 = self._get_max_and_intervals(self.sampler.flatchain[:,1])
logMacc1, logMacc, logMacc2 = self._get_max_and_intervals(self.sampler.flatchain[:,2])
R_out1, R_out, R_out2 = self._get_max_and_intervals(self.sampler.flatchain[:,3])
#scale1, scale, scale2 = self._get_max_and_intervals(self.sampler.flatchain[:,3])
self.Mstar = Mstar
self.Mstarerr1 = Mstar - Mstar1
self.Mstarerr2 = Mstar2 - Mstar
self.Rstar = Rstar
self.Rstarerr1 = Rstar - Rstar1
self.Rstarerr2 = Rstar2 - Rstar
self.logMacc = logMacc
self.logMaccerr1 = logMacc - logMacc1
self.logMaccerr2 = logMacc2 - logMacc
self.R_out = R_out
self.R_outerr1 = R_out - R_out1
self.R_outerr2 = R_out2 - R_out
def _save_output(self):
'''
Saves in a results file.
'''
exists = os.path.isfile(self.resfile)
with open(self.resfile, 'a') as outfile:
print ("Saving results to %s"%self.resfile)
if self.model == "BlackBody":
if not exists:
outfile.write("mjd T Terr1 Terr2 R Rerr1 Rerr2 L Lerr1 Lerr2 Av_MW\n")
outfile.write("%.5f %.3f %.3f %.3f %.3f %.3f %.3f %.3e %.3e %.3e %.3f\n"%\
(self.mjd, self.T, self.Terr1, self.Terr2, self.R, self.Rerr1, self.Rerr2, self.L, self.Lerr1, self.Lerr2, self.av_mw))
elif self.model == "BlackBody_Av":
if not exists:
outfile.write("mjd T Terr1 Terr2 R Rerr1 Rerr2 L Lerr1 Lerr2 Av Averr1 Averr2 Av_MW\n")
outfile.write("%.5f %.3f %.3f %.3f %.3f %.3f %.3f %.3e %.3e %.3e %.3f %.3f %.3f %.3f\n"%\
(self.mjd, self.T, self.Terr1, self.Terr2, self.R, self.Rerr1, self.Rerr2, \
self.L, self.Lerr1, self.Lerr2, self.Av, self.Averr1, self.Averr2, self.av_mw))
elif self.model == "BlackBody2":
if not exists:
outfile.write("mjd T Terr1 Terr2 R Rerr1 Rerr2 L Lerr1 Lerr2 Tsec Tsecerr1 Tsecerr2 Rsec Rsecerr1 Rsecerr2 Lsec Lsecerr1 Lsecerr2 Av_MW\n")
outfile.write("%.5f %.3f %.3f %.3f %.3f %.3f %.3f %.3e %.3e %.3e %.3f %.3f %.3f %.3f %.3f %.3f %.3e %.3e %.3e %.3f \n"%\
(self.mjd, self.T, self.Terr1, self.Terr2, self.R, self.Rerr1, self.Rerr2, \
self.L, self.Lerr1, self.Lerr2,
self.Tsec, self.Tsecerr1, self.Tsecerr2, self.Rsec, self.Rsecerr1, self.Rsecerr2, \
self.Lsec, self.Lsecerr1, self.Lsecerr2, self.av_mw))
elif self.model == "BlackBody2_Av":
if not exists:
outfile.write("mjd T Terr1 Terr2 R Rerr1 Rerr2 L Lerr1 Lerr2 Av Averr1 Averr2 Tsec Tsecerr1 Tsecerr2 Rsec Rsecerr1 Rsecerr2 Av_MW\n")
outfile.write("%.5f %.3f %.3f %.3f %.3f %.3f %.3f %.3e %.3e %.3e %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f\n"%\
(self.mjd, self.T, self.Terr1, self.Terr2, self.R, self.Rerr1, self.Rerr2, \
self.L, self.Lerr1, self.Lerr2, self.Av, self.Averr1, self.Averr2,\
self.Tsec, self.Tsecerr1, self.Tsecerr2, self.Rsec, self.Rsecerr1, self.Rsecerr2, self.av_mw))
elif self.model == "PowerLaw":
if not exists:
outfile.write("mjd alpha alphaerr1 alphaerr2 scale scaleerr1 scaleerr2 Av Averr1 Averr2 Av_MW\n")
outfile.write("%.5f %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f\n"%\
(self.mjd, self.alpha, self.alphaerr1, self.alphaerr2, self.scale, self.scaleerr1, self.scaleerr2, \
self.Av, self.Averr1, self.Averr2, self.av_mw))
elif self.model == "PowerLaw_BlackBody":
if not exists:
outfile.write("mjd alpha alphaerr1 alphaerr2 scale scaleerr1 scaleerr2 T Terr1 Terr2 R Rerr1 Rerr2 Av_MW\n")
outfile.write("%.5f %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f %.3f\n"%\
(self.mjd, self.alpha, self.alphaerr1, self.alphaerr2, self.scale, self.scaleerr1, self.scaleerr2, \
self.T, self.Terr1, self.Terr2, \
self.R, self.Rerr1, self.Rerr2, \
self.av_mw))
elif self.model == "Disk":
if not exists:
outfile.write("mjd M Merr1 Merr2 Rstar Rerr1 Rerr2 Macc Maccerr1 Maccerr2 R_out R_outerr1 R_outerr2 Av_MW\n")
outfile.write("%.5f %.3f %.3f %.3f %.3f %.3f %.3f %.3e %.3e %.3e %.3e %.3e %.3e %.3f\n"%\
(self.mjd, self.Mstar, self.Mstarerr1, self.Mstarerr1, \
self.Rstar, self.Rstarerr1, self.Rstarerr2,\
self.logMacc, self.logMaccerr1, self.logMaccerr2,\
#self.scale, self.scaleerr1, self.scaleerr2, \
self.R_out, self.R_outerr1, self.R_outerr2,\
self.av_mw))
else:
print ("Unknown model! %s"%self.model)
def _get_bol_lum(self, T, R):
'''
T is in K
R in R_sun.
Gives the Lbol in Lsun
'''
L = cnt.sigma_sb * (T * u.K)**4 * 4 * np.pi * (R*u.Rsun)**2
return (L.to(u.Lsun)).value
def _get_save_path(self, savefile, plot_name=""):
'''
Checks what savefile name has been given.
If there is a value, then it jsut stores it in the plot directory provided.
If there is no name, then it creates a filename with the suffix provided.
It also checks if there is already a file named like that, and it that is the case,
it increases the suffix so that it has a higher number, avoiding collision.
'''
#If there is a given name to store the file, then we use that one
if (not savefile is None):
if os.path.dirname(savefile) == "":
name = os.path.join(self.plotdir, os.path.basename(savefile))
#If there is no name, then we will save the plots in the plot directory
#with an automatic name.
# This name will increase a count if the name exists already.
else:
i = 0
name = os.path.join(self.plotdir, "%s_%.1f_%d.pdf"%(plot_name, self.mjd, i))
while (os.path.isfile(name)):
i = i+1
name = os.path.join(self.plotdir, "%s_%.1f_%d.pdf"%(plot_name, self.mjd, i))
return name
def _initialize_parameters(self, plot=False):
'''
Runs the least squares optimiztion routine to find the best initial parameters
to start the MCMC with.
'''
lam = np.linspace(np.min(self.wls)*0.9, np.max(self.wls)*1.1, 2000)
a_v_wls = extinction.fitzpatrick99(self.wls, a_v=self.av_mw, unit='aa')
reddening = 10**(0.4*a_v_wls)
if self.model == "BlackBody":
flux_ini = self._model_2(lam, self.initT1, self.initR1)
p0 = (self.initT1, self.initR1)
print ("Initial parameters given:", p0)
#Perform a LSQ fit
#params, covar = curve_fit(self._model_2, self.wls , self.fluxes, \
#p0 = p0, sigma=self.fluxerrs, absolute_sigma=True, maxfev = 20000)
#flux_end = self._model_2(lam, *params)
if plot:
plt.clf()
mask_lims = self.fluxerrs<0
plt.plot(lam, flux_ini, "r--", label="Fit initial parameters")
#plt.plot(lam, flux_end, label="Best fit LSQ")
plt.errorbar(self.wls[~mask_lims], self.fluxes[~mask_lims], yerr=self.fluxerrs[~mask_lims], marker="o", color="b", lw=0, label="Measurements")
plt.errorbar(self.wls[mask_lims], self.fluxes[mask_lims], yerr=self.fluxes[mask_lims]*0.2, fmt="o", color="b", uplims=True)
plt.xlabel("Wavelength [A]")
plt.ylabel("$F_{\\lambda}$ [erg/s/cm2/A]")
plt.ylim(0.8*np.min(self.fluxes), 1.2*np.max(self.fluxes))
plt.legend()
plt.yscale("log")
name = self._get_save_path(None, "fluxes_obs_bb")
plt.savefig(name, dpi=200)
elif self.model == "BlackBody_Av":
flux_ini = self._model_av_r_2(lam, self.initT1, self.initR1, self.av_host)
p0 = (self.initT1, self.initR1, self.av_host)
print ("Initial ", p0)
#params, covar = curve_fit(self._model_av_r_2, self.wls , self.fluxes, \
#p0 = p0, sigma=self.fluxerrs, absolute_sigma=True, maxfev = 20000)
#flux_end = self._model_av_r_2(lam, *params)
if plot:
plt.clf()
plt.plot(lam, flux_ini, "r--", label="Fit initial parameters")
#plt.plot(lam, flux_end, label="Best fit LSQ")
plt.errorbar(self.wls, self.fluxes, yerr=self.fluxerrs, marker="o", lw=0, label="Measurements")
plt.xlabel("Wavelength [A]")
plt.ylabel("$L_{\\lambda}$ [erg/s/A]")
plt.ylim(0.8*np.min(self.fluxes), 1.2*np.max(self.fluxes))
plt.legend()
name = self._get_save_path(None, "fluxes_obs_bb_av")
plt.savefig(name, dpi=200)
elif self.model == "BlackBody2_Av":
flux_ini = self._model2_av_r_2(lam, self.initT1, self.initR1, self.av_host, self.initT2, self.initR2)
p0 = (self.initT1, self.initR1, self.av_host, self.initT2, self.initR2)
print ("Initial ", p0)
#params, covar = curve_fit(self._model2_av_r_2, self.wls , self.fluxes, \
#p0 = p0, sigma=self.fluxerrs, absolute_sigma=True, maxfev = 20000)
#flux_end = self._model2_av_r_2(lam, *params)
if plot:
plt.clf()
plt.plot(lam, flux_ini, "r--", label="Fit initial parameters")
#plt.plot(lam, flux_end, label="Best fit LSQ")
plt.errorbar(self.wls, self.fluxes, yerr=self.fluxerrs, marker="o", lw=0, label="Measurements")
plt.xlabel("Wavelength [A]")
plt.ylabel("$L_{\\lambda}$ [erg/s/A]")
plt.ylim(0.8*np.min(self.fluxes), 1.2*np.max(self.fluxes))
plt.legend()
name = self._get_save_path(None, "fluxes_obs")
plt.savefig(name, dpi=200)
elif self.model == "BlackBody2":
flux_ini = self._model2_r_2(lam, self.initT1, self.initR1, self.initT2, self.initR2)
p0 = (self.initT1, self.initR1, self.initT2, self.initR2)
print ("Initial ", p0)
#params, covar = curve_fit(self._model2_r_2, self.wls , self.fluxes, \
#p0 = p0, sigma=self.fluxerrs, absolute_sigma=True, maxfev = 20000)
#flux_end = self._model2_r_2(lam, *params)
#flux_1 = self._model_2(lam, *params[0:2])
#flux_2 = self._model_2(lam, *params[2:])
if plot:
plt.clf()
plt.figure(figsize=(6,4))
plt.plot(lam, flux_ini, "r--", label="Fit initial parameters")
#plt.plot(lam, flux_end, label="Best fit LSQ")
#plt.plot(lam, flux_1, label="BB1")
#plt.plot(lam, flux_2, label="BB2")
plt.errorbar(self.wls, self.fluxes, yerr=self.fluxerrs, marker="o", lw=0, label="Measurements")
plt.xlabel("Wavelength [A]")
plt.ylabel("$L_{\\lambda}$ [erg/s/A]")
plt.legend(loc="best", fontsize=10)
plt.ylim(0.8*np.min(self.fluxes), 1.2*np.max(self.fluxes))
plt.yscale("log")
name = self._get_save_path(None, "fluxes_obs_2bb")
plt.savefig(name, dpi=200)
elif self.model == "PowerLaw":
#params, covar = curve_fit(self._model_powerlaw_2, self.wls , self.fluxes, \
#p0=(self.alpha, self.initR1, self.av_host), sigma=self.fluxerrs, absolute_sigma=True, maxfev = 10000)
lam = np.linspace(3000, 25000, 2000)
fluxpw = self._model_powerlaw_2(lam, self.alpha, self.scale, self.av_host)
if plot:
plt.clf()
plt.plot(lam, fluxpw, label="Fit initial parameters")
plt.errorbar(self.wls, self.fluxes, yerr=self.fluxerrs, marker="o", lw=0, label="Measurements")
plt.xlabel("Wavelength [A]")
plt.ylabel("$L_{\\lambda}$ [erg/s/A]")
plt.ylim(0.8*np.min(self.fluxes), 1.2*np.max(self.fluxes))
plt.legend()
name = self._get_save_path(None, "fluxes_obs_powerlaw")
plt.savefig(name, dpi=200)
print ("Saved fit as %s"%name)
elif self.model == "PowerLaw_BlackBody":
#params, covar = curve_fit(self._model_powerlaw_2, self.wls , self.fluxes, \
#p0=(self.alpha, self.initR1, self.av_host), sigma=self.fluxerrs, absolute_sigma=True, maxfev = 10000)
lam = np.linspace(3000, 25000, 2000)
fluxpw = self._model_powerlaw_bb_2(lam, self.alpha, self.scale, self.initT1, self.initR1)
if plot:
plt.clf()
plt.plot(lam, fluxpw, label="Fit initial parameters")
plt.errorbar(self.wls, self.fluxes, yerr=self.fluxerrs, marker="o", lw=0, label="MW ext. corr")
plt.errorbar(self.wls, self.fluxes/reddening, yerr=self.fluxerrs, marker="o", lw=0, label="Measurements")
plt.xlabel("Wavelength [A]")
plt.ylabel("$L_{\\lambda}$ [erg/s/A]")
plt.ylim(0.8*np.min(self.fluxes/reddening), 1.2*np.max(self.fluxes))
plt.legend(loc="best")
name = self._get_save_path(None, "fluxes_obs_powerlaw_bb")
plt.savefig(name, dpi=200)
print ("Saved fit as %s"%name)
if self.model == 'Disk':
#params = (0.5, 0.2, 5e-9, 1, 2)
p0 = (self.Mstar, self.Rstar, self.logMacc, self.R_out)
#params, covar = curve_fit(self._model_accretion_disk2, self.wls , self.fluxes, \
#p0 = p0, sigma=self.fluxerrs, absolute_sigma=True, maxfev = 20000)
#print ("LSQ fit: Mstar:", params[0], " Rstar", params[1], "logMacc ", \
# params[2], "R_out", params[3])
lam = np.linspace(3000, 25000, 2000)
#flux_disk = self._model_accretion_disk2(lam, params[0], params[1], params[2], params[3])
if plot:
plt.clf()
plt.errorbar(self.wls, self.fluxes, yerr=self.fluxerrs, marker="o", lw=0, label="Measurements")
#plt.plot(lam, flux_disk, lw=3)
plt.xlabel("Wavelength [$\\mu$m]")
plt.ylabel("Flux [erg/cm$^2$/s]")
plt.ylim(np.nanmin(self.fluxes)*0.9, np.nanmax(self.fluxes)*1.2)
plt.legend()
name = self._get_save_path(None, "fluxes_obs_disk")
plt.savefig(name, dpi=200)
print ("Saved fit as %s"%name)
def initialize(self, plot=False):
'''
Will transform the magnitudes to fluxes and use the distance to the object to
calculate the luminosity at each wavelength.
'''
if (not os.path.isdir(self.plotdir)):
os.makedirs(self.plotdir)
print ("Created plot directory %s"%self.plotdir)
#Directory where to store the results
if (not os.path.isdir(self.resdir)):
os.makedirs(self.resdir)
print ("Created result directory %s"%(self.resdir))
self.resfile = os.path.join(self.resdir, self.model + os.path.basename(self.resfile))
# generate the data
with warnings.catch_warnings():
warnings.simplefilter("ignore")
self.wls, self.fluxes, self.fluxerrs = self._band2flux()
#Plot the raw fluxes before correcting them.
'''if (plot):
plt.figure(figsize=(8,6))
plt.errorbar(self.wls, self.fluxes, yerr=self.fluxerrs, marker="o", lw=0)
for i in range(len(self.wls)):
plt.text(self.wls[i], self.fluxes[i]*1.01, self.bands[i].split(",")[-1], alpha=.4)
name = self._get_save_path(None, "fluxes_observed")
plt.yscale("log")
plt.xlabel("Wavelength [A]")
plt.ylabel("log (Flux/[erg/cm2/s])")
plt.tight_layout()
plt.savefig(name, dpi=200)'''
if not self.distMpc is None and self.distMpc !=0:
print ("Using distance to the source of %.1e Mpc"%self.distMpc)
fluxFactor = (4*np.pi*((self.distMpc*u.Mpc).to(u.cm) )**2).value
elif (self.distMpc is None or self.distMpc==0 )and (not self.z is None and self.z != 0):
self.distMpc = self.cosmo.luminosity_distance(self.z)
#Compute the flux multiplication factor for the object if it is at distance distMpc
#We transform that to cm, as the flux is in erg cm-2 s-1
fluxFactor = (4*np.pi*(self.distMpc.to(u.cm) )**2).value
else: # self.distMpc is None and self.z is None:
#Here we do not use any multiplication flux factor
print ("Warning: no redshift or distance provided!")
fluxFactor = 1
self.fluxes = self.fluxes * fluxFactor
self.fluxerrs = self.fluxerrs * fluxFactor
self._initialize_parameters(plot)
def run(self):
'''
Runs the main MCMC process.
Retrieves the priors, the likelihood process and computes the posterior probability.
'''
xs = self.wls
ys = self.fluxes
errs = self.fluxerrs
if self.model == "BlackBody":
p0 = np.array([ self.initT1, self.initR1])
sigs = np.array([self.initT1*0.2, self.initR1*0.2])
elif self.model == "BlackBody_Av":
p0 = np.array([ self.initT1, self.initR1, self.av_host])
sigs = np.array([2000, 10, 0.5])
elif self.model == "BlackBody2":
p0 = np.array([ self.initT1, self.initR1, self.initT2, self.initR2])
sigs = np.array([self.initT1*0.2, self.initR1*0.2, self.initT2*0.2, self.initR2*0.2])
elif self.model == "BlackBody2_Av":
p0 = np.array([ self.initT1, self.initR1, self.av_host, self.initT2, self.initR2])
sigs = np.array([2000, 5, 1, 2000, 5])
elif self.model == "PowerLaw":
p0 = np.array([ self.alpha, self.scale, self.av_host])
sigs = np.array([2, 3, 2])
elif self.model == "PowerLaw_BlackBody":
p0 = np.array([ self.alpha, self.scale, self.initT1, self.initR1])
sigs = np.array([2, 3, 2000, 2])
elif self.model == "Disk":
p0 = np.array([ self.Mstar, self.Rstar, self.logMacc, self.R_out])
sigs = np.array([0.1, 0.01, 1, 0.1])
print ("Initialized with p0", p0, " and sigmas ", sigs)
else:
print ("-------------------CRITICAL ERROR!----------------------")
print ("-------------------UNKNOWN model! %s----------------------"%self.model)
print ("-------------------CRITICAL ERROR!----------------------")
sys.exit()
ndim = len(p0)
# emsemble MCMC
p0s = emcee.utils.sample_ball(p0, sigs, self.nwalkers)
# initialize the ball of initial conditions
#Supports the threads=X argument for parallelization
sampler = emcee.EnsembleSampler(self.nwalkers, ndim, self._logposterior,\
args=(xs, ys, errs), threads=10)
pos, lnprob, state = sampler.run_mcmc(p0s, self.burnin)
print ("Burning phase finished")
sampler.reset()
pos, lnprob, state = sampler.run_mcmc(pos, self.niterations)
print ('Acceptance ratio', sampler.acceptance_fraction)
self.sampler = sampler
print ("MCMC main phase finished")
self._fill_output()
self._save_output()
def plot_corner_posteriors(self, savefile=None):
'''
Plots the corner plot of the MCMC results.
'''
if self.model == "BlackBody2":
labels=["T1", "R1", "T2", "R2"]
elif self.model.startswith("BlackBody"):
labels=["T1", "R1", "Av", "T2", "R2"]
elif self.model == "PowerLaw":
labels=["alpha", "scale", "Av"]
elif self.model == "PowerLaw_BlackBody":
labels = ["alpha", "scale", "T", "R"]
elif self.model == "Disk":
labels = ["Mstar", "Rstar", "logMacc", "R_out"]
ndim = len(self.sampler.flatchain[0,:])
chain = self.sampler
samples = chain.flatchain
samples = samples[:,0:ndim]
plt.figure(figsize=(8,8))
fig = corner.corner(samples, labels=labels[0:ndim], quantiles=[0.16, 0.5, 0.84],
show_titles=True, title_kwargs={"fontsize": 12})
fig.suptitle("MJD: %.2f"%self.mjd)
name = self._get_save_path(savefile, "mcmc_posteriors")
plt.savefig(name)
plt.close("all")
plt.figure(figsize=(8,ndim*3))
for n in range(ndim):
plt.subplot(ndim,1,n+1)
chain = self.sampler.chain[:,:,n]
nwalk, nit = chain.shape
for i in np.arange(nwalk):
plt.plot(chain[i], lw=0.1)
plt.ylabel(labels[n])
plt.xlabel("Iteration")
name_walkers = self._get_save_path(savefile, "mcmc_walkers")
plt.tight_layout()
plt.savefig(name_walkers)
plt.close("all")
def plot_fit(self, lambdaFlambda=False):
'''
Plots the best fit model to the data.
'''
lam = np.linspace( np.min(self.wls) -1500 , np.max(self.wls) + 1500, 1000)
plt.clf()
plt.figure(figsize=(8,6))
mask_lims = self.fluxerrs<0
if lambdaFlambda:
factor_obs=self.wls
else:
factor_obs=np.ones_like(self.wls)
plt.errorbar(self.wls[~mask_lims], self.fluxes[~mask_lims]*factor_obs[~mask_lims], yerr=self.fluxerrs[~mask_lims]*factor_obs[~mask_lims], marker="o", color="b", lw=0, label="Measurements")
plt.errorbar(self.wls[mask_lims], self.fluxes[mask_lims]*factor_obs[mask_lims], yerr=self.fluxes[mask_lims]*0.2*factor_obs[mask_lims], fmt="o", color="b", uplims=True)
for i in range(len(self.wls)):
plt.text(self.wls[i], self.fluxes[i]*1.01*factor_obs[i], self.bands[i], alpha=.4, fontsize=8)
if self.model == "BlackBody":
fluxbb = self._model(lam, (self.T, self.R))
if lambdaFlambda:
factor = lam
else:
factor = np.ones_like(lam)
plt.plot(lam, fluxbb*factor, "k-", label="BB fit")
plt.title("T: %d K R:%d R$_{\odot}$ Lumiosity %.2e L$_{\odot}$"%(self.T, self.R, self.L))
elif self.model == "BlackBody_Av":
fluxbb = self._model(lam, (self.T, self.R))
fluxbb_red = self._model_av_r(lam, (self.T, self.R, self.Av))
plt.plot(lam, fluxbb, "k-", label="BB fit")
plt.plot(lam, fluxbb_red, "red", label="BB fit + reddening")
plt.title("T: %.1f K R:%.1f R$_{\odot}$ Lumiosity %.1e L$_{\odot}$ Av: %.2f"%(np.round(self.T,0), np.round(self.R,0), np.round(self.L,1), self.Av))
elif self.model == "BlackBody2_Av":
fluxbb_red = self._model2_av(lam, (self.T, self.R, self.Av))
fluxbb_secondary_red = self._model2_av(lam, (self.Tsec, self.Rsec, self.Av))
fluxbb_with_seconday = self._model2_av(lam, (self.T, self.R, self.Av, self.Tsec, self.Rsec))
plt.plot(lam, fluxbb_red, "k-", label="BB1 fit + reddening")
plt.plot(lam, fluxbb_secondary_red, "k--", label="BB2 fit + reddening")
plt.plot(lam, fluxbb_with_seconday, "green", label="BB1 + BB2")
plt.title("T: %.1f K R:%.1f R$_{\odot}$ Lumiosity %.1e L$_{\odot}$ Av: %.2f\n T2: %.1f R2: %.1f"%(self.T, \
self.R, self.L, self.Av, self.Tsec, self.Rsec))
elif self.model == "BlackBody2":
fluxbb_primary = self._model(lam, (self.T, self.R))
fluxbb_secondary = self._model(lam, (self.Tsec, self.Rsec))
fluxbb_with_seconday = self._model2_r(lam, (self.T, self.R, self.Tsec, self.Rsec))
plt.plot(lam, fluxbb_primary, "k-", label="BB1 fit")
plt.plot(lam, fluxbb_secondary, "k--", label="BB2 fit")
plt.plot(lam, fluxbb_with_seconday, "green", label="BB1 + BB2")
plt.title("T: %d K R:%d R$_{\odot}$ T2: %d R2: %d"%( self.T, \
self.R, self.Tsec, self.Rsec))
elif self.model == "PowerLaw":
flux = self._model_powerlaw(lam, (self.alpha, self.scale, self.Av))
plt.plot(lam, flux, "k-", label="PowerLaw + reddening")
plt.title("$\\alpha$: %.1f Av: %.2f"%(self.alpha, self.Av))
elif self.model == "PowerLaw_BlackBody":
flux = self._model_powerlaw_bb(lam, (self.alpha, self.scale, self.T, self.R))
flux_pw = self._model_powerlaw(lam, (self.alpha, self.scale, 0))
flux_bb = self._model(lam, (self.T, self.R))
plt.plot(lam, flux, "k-", label="PowerLaw + BlackBody")
plt.plot(lam, flux_pw, "b--", label="PowerLaw")
plt.plot(lam, flux_bb, "g:", label="BlackBody")
plt.title("$\\alpha$: %.1f scale: %.2e T: %.1f R:%.1f"%(self.alpha, self.scale, self.T, self.R))
elif self.model == "Disk":
fluxdisk = self._model_accretion_disk(lam, (self.Mstar, self.Rstar, self.logMacc, self.R_out))
plt.plot(lam, fluxdisk, "k-", label="Disk fit")
plt.title("M:%.3f M$_{\\odot}$ R:%.3f R$_{\odot}$ M$_{acc}$:%.2f R_out: %.2f"%(self.Mstar, self.Rstar, self.logMacc, self.R_out))
ymin, ymax = plt.ylim()
#plt.ylim(np.max([ymin, np.min(self.fluxes)*0.01]), ymax)
plt.xlabel("Wavelength [$\\AA$]")
if (lambdaFlambda):
plt.ylabel("$\\lambda F_{\\lambda}$ [erg/s]")
plt.ylim(ymin=np.min(self.fluxes*factor_obs) * 0.1)
else:
plt.ylabel("$F_{\\lambda}$ [erg/s/$\\AA$]")
plt.ylim(ymin=np.min(self.fluxes) * 0.1)
plt.yscale("log")
plt.legend()
name = self._get_save_path(None, "mcmc_best_fit_model")
plt.savefig(name)
plt.close("all")
def write_fit_params(self):
'''
Write the best fit parameters of the model to the standard output.
'''
if self.model.startswith("BlackBody"):
#Prints the best parameters
print ('''
Temperature: \t %.3f -%.3f +%.3f K
Radius: \t\t %.2e -%.2e +%.2e R$_{\odot}$
Luminosity: \t %.3e -%.3e +%.3e L$_{\odot}$'''%(\
self.T, self.Terr1, self.Terr2, \
self.R, self.Rerr1, self.Rerr2, \
self.L, self.Lerr1, self.Lerr2))
if self.model == "BlackBody_Av":
print (" Av: \t\t\t %.1f -%.1f +%.1f mag"%(self.Av, self.Averr1, self.Averr2))
if self.model == "BlackBody2":
print (" Temperature2: %.1f -%.1f +%.1f K"%(self.Tsec, self.Tsecerr1, self.Tsecerr2))
print (" Radius2: %.2e -%.2e +%.2e R$_{\odot}$"%(self.Rsec, self.Rsecerr1, self.Rsecerr2))
print (" Luminosity2 %.3e -%.3e +%.3e L$_{\odot}$"%(self.Lsec, self.Lsecerr1, self.Lsecerr2))
if self.model == "BlackBody2_Av":
print (" Av: %.1f -%.1f +%.1f mag"%(self.Av, self.Averr1, self.Averr2))
print (" Temperature2: %.1f -%.1f +%.1f K"%(self.Tsec, self.Tsecerr1, self.Tsecerr2))
print (" Radius2: %.1f -%.1f +%.1f R$_{\odot}$"%(self.Rsec, self.Rsecerr1, self.Rsecerr2))
if (self.model == "PowerLaw"):
print ('''
alpha: %.2f -%.2f +%.2f
Scale : %.2e -%.2e +%.2e
Av %.2f -%.2f +%.2f'''%(\
self.alpha, self.alphaerr1, self.alphaerr2, \
self.scale, self.scaleerr1, self.scaleerr2, \
self.Av, self.Averr1, self.Averr2))
if (self.model == "PowerLaw_BlackBody"):
print ('''
alpha: %.2f -%.2f +%.2f
Scale (R): %.2e -%.2e +%.2e
T %.2f -%.2f +%.2f
R %.2f -%.2f +%.2f '''%(\
self.alpha, self.alphaerr1, self.alphaerr2, \
self.scale, self.scaleerr1, self.scaleerr2,\
self.T, self.Terr1, self.Terr2,\
self.R, self.Rerr1, self.Rerr2 ))
if (self.model == "Disk"):
print ('''
Mstar: %.3f$_{-%.3f}^{+%.3f}$
Rstar (10^8 cm): %.3f -%.3f +%.3f
logMacc %.3f$_{-%.3f}^{+%.3f}$
R_out %.3f$_{-%.3f}^{+%.3f}$ '''%(\
self.Mstar, self.Mstarerr1, self.Mstarerr2, \
self.Rstar*(u.Rsun.to(u.cm))/1e8, self.Rstarerr1*(u.Rsun.to(u.cm))/1e8, self.Rstarerr2*(u.Rsun.to(u.cm))/1e8,\
self.logMacc, self.logMaccerr1, self.logMaccerr2,\
self.R_out, self.R_outerr1, self.R_outerr2 ))
| mit |
Jericho/deep-learning | image-classification/helper.py | 155 | 5631 | import pickle
import numpy as np
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelBinarizer
def _load_label_names():
"""
Load the label names from file
"""
return ['airplane', 'automobile', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
def load_cfar10_batch(cifar10_dataset_folder_path, batch_id):
"""
Load a batch of the dataset
"""
with open(cifar10_dataset_folder_path + '/data_batch_' + str(batch_id), mode='rb') as file:
batch = pickle.load(file, encoding='latin1')
features = batch['data'].reshape((len(batch['data']), 3, 32, 32)).transpose(0, 2, 3, 1)
labels = batch['labels']
return features, labels
def display_stats(cifar10_dataset_folder_path, batch_id, sample_id):
"""
Display Stats of the the dataset
"""
batch_ids = list(range(1, 6))
if batch_id not in batch_ids:
print('Batch Id out of Range. Possible Batch Ids: {}'.format(batch_ids))
return None
features, labels = load_cfar10_batch(cifar10_dataset_folder_path, batch_id)
if not (0 <= sample_id < len(features)):
print('{} samples in batch {}. {} is out of range.'.format(len(features), batch_id, sample_id))
return None
print('\nStats of batch {}:'.format(batch_id))
print('Samples: {}'.format(len(features)))
print('Label Counts: {}'.format(dict(zip(*np.unique(labels, return_counts=True)))))
print('First 20 Labels: {}'.format(labels[:20]))
sample_image = features[sample_id]
sample_label = labels[sample_id]
label_names = _load_label_names()
print('\nExample of Image {}:'.format(sample_id))
print('Image - Min Value: {} Max Value: {}'.format(sample_image.min(), sample_image.max()))
print('Image - Shape: {}'.format(sample_image.shape))
print('Label - Label Id: {} Name: {}'.format(sample_label, label_names[sample_label]))
plt.axis('off')
plt.imshow(sample_image)
def _preprocess_and_save(normalize, one_hot_encode, features, labels, filename):
"""
Preprocess data and save it to file
"""
features = normalize(features)
labels = one_hot_encode(labels)
pickle.dump((features, labels), open(filename, 'wb'))
def preprocess_and_save_data(cifar10_dataset_folder_path, normalize, one_hot_encode):
"""
Preprocess Training and Validation Data
"""
n_batches = 5
valid_features = []
valid_labels = []
for batch_i in range(1, n_batches + 1):
features, labels = load_cfar10_batch(cifar10_dataset_folder_path, batch_i)
validation_count = int(len(features) * 0.1)
# Prprocess and save a batch of training data
_preprocess_and_save(
normalize,
one_hot_encode,
features[:-validation_count],
labels[:-validation_count],
'preprocess_batch_' + str(batch_i) + '.p')
# Use a portion of training batch for validation
valid_features.extend(features[-validation_count:])
valid_labels.extend(labels[-validation_count:])
# Preprocess and Save all validation data
_preprocess_and_save(
normalize,
one_hot_encode,
np.array(valid_features),
np.array(valid_labels),
'preprocess_validation.p')
with open(cifar10_dataset_folder_path + '/test_batch', mode='rb') as file:
batch = pickle.load(file, encoding='latin1')
# load the test data
test_features = batch['data'].reshape((len(batch['data']), 3, 32, 32)).transpose(0, 2, 3, 1)
test_labels = batch['labels']
# Preprocess and Save all test data
_preprocess_and_save(
normalize,
one_hot_encode,
np.array(test_features),
np.array(test_labels),
'preprocess_test.p')
def batch_features_labels(features, labels, batch_size):
"""
Split features and labels into batches
"""
for start in range(0, len(features), batch_size):
end = min(start + batch_size, len(features))
yield features[start:end], labels[start:end]
def load_preprocess_training_batch(batch_id, batch_size):
"""
Load the Preprocessed Training data and return them in batches of <batch_size> or less
"""
filename = 'preprocess_batch_' + str(batch_id) + '.p'
features, labels = pickle.load(open(filename, mode='rb'))
# Return the training data in batches of size <batch_size> or less
return batch_features_labels(features, labels, batch_size)
def display_image_predictions(features, labels, predictions):
n_classes = 10
label_names = _load_label_names()
label_binarizer = LabelBinarizer()
label_binarizer.fit(range(n_classes))
label_ids = label_binarizer.inverse_transform(np.array(labels))
fig, axies = plt.subplots(nrows=4, ncols=2)
fig.tight_layout()
fig.suptitle('Softmax Predictions', fontsize=20, y=1.1)
n_predictions = 3
margin = 0.05
ind = np.arange(n_predictions)
width = (1. - 2. * margin) / n_predictions
for image_i, (feature, label_id, pred_indicies, pred_values) in enumerate(zip(features, label_ids, predictions.indices, predictions.values)):
pred_names = [label_names[pred_i] for pred_i in pred_indicies]
correct_name = label_names[label_id]
axies[image_i][0].imshow(feature)
axies[image_i][0].set_title(correct_name)
axies[image_i][0].set_axis_off()
axies[image_i][1].barh(ind + margin, pred_values[::-1], width)
axies[image_i][1].set_yticks(ind + margin)
axies[image_i][1].set_yticklabels(pred_names[::-1])
axies[image_i][1].set_xticks([0, 0.5, 1.0])
| mit |
xiaoxiamii/scikit-learn | examples/neighbors/plot_digits_kde_sampling.py | 251 | 2022 | """
=========================
Kernel Density Estimation
=========================
This example shows how kernel density estimation (KDE), a powerful
non-parametric density estimation technique, can be used to learn
a generative model for a dataset. With this generative model in place,
new samples can be drawn. These new samples reflect the underlying model
of the data.
"""
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import load_digits
from sklearn.neighbors import KernelDensity
from sklearn.decomposition import PCA
from sklearn.grid_search import GridSearchCV
# load the data
digits = load_digits()
data = digits.data
# project the 64-dimensional data to a lower dimension
pca = PCA(n_components=15, whiten=False)
data = pca.fit_transform(digits.data)
# use grid search cross-validation to optimize the bandwidth
params = {'bandwidth': np.logspace(-1, 1, 20)}
grid = GridSearchCV(KernelDensity(), params)
grid.fit(data)
print("best bandwidth: {0}".format(grid.best_estimator_.bandwidth))
# use the best estimator to compute the kernel density estimate
kde = grid.best_estimator_
# sample 44 new points from the data
new_data = kde.sample(44, random_state=0)
new_data = pca.inverse_transform(new_data)
# turn data into a 4x11 grid
new_data = new_data.reshape((4, 11, -1))
real_data = digits.data[:44].reshape((4, 11, -1))
# plot real digits and resampled digits
fig, ax = plt.subplots(9, 11, subplot_kw=dict(xticks=[], yticks=[]))
for j in range(11):
ax[4, j].set_visible(False)
for i in range(4):
im = ax[i, j].imshow(real_data[i, j].reshape((8, 8)),
cmap=plt.cm.binary, interpolation='nearest')
im.set_clim(0, 16)
im = ax[i + 5, j].imshow(new_data[i, j].reshape((8, 8)),
cmap=plt.cm.binary, interpolation='nearest')
im.set_clim(0, 16)
ax[0, 5].set_title('Selection from the input data')
ax[5, 5].set_title('"New" digits drawn from the kernel density model')
plt.show()
| bsd-3-clause |
mwv/scikit-learn | sklearn/preprocessing/__init__.py | 268 | 1319 | """
The :mod:`sklearn.preprocessing` module includes scaling, centering,
normalization, binarization and imputation methods.
"""
from ._function_transformer import FunctionTransformer
from .data import Binarizer
from .data import KernelCenterer
from .data import MinMaxScaler
from .data import MaxAbsScaler
from .data import Normalizer
from .data import RobustScaler
from .data import StandardScaler
from .data import add_dummy_feature
from .data import binarize
from .data import normalize
from .data import scale
from .data import robust_scale
from .data import maxabs_scale
from .data import minmax_scale
from .data import OneHotEncoder
from .data import PolynomialFeatures
from .label import label_binarize
from .label import LabelBinarizer
from .label import LabelEncoder
from .label import MultiLabelBinarizer
from .imputation import Imputer
__all__ = [
'Binarizer',
'FunctionTransformer',
'Imputer',
'KernelCenterer',
'LabelBinarizer',
'LabelEncoder',
'MultiLabelBinarizer',
'MinMaxScaler',
'MaxAbsScaler',
'Normalizer',
'OneHotEncoder',
'RobustScaler',
'StandardScaler',
'add_dummy_feature',
'PolynomialFeatures',
'binarize',
'normalize',
'scale',
'robust_scale',
'maxabs_scale',
'minmax_scale',
'label_binarize',
]
| bsd-3-clause |
tdegeus/GooseEYE | docs/examples/clusters_dilate_periodic.py | 1 | 2926 | r'''
Plot and/or check.
Usage:
script [options]
Options:
-s, --save Save output for later check.
-c, --check Check against earlier results.
-p, --plot Plot.
-h, --help Show this help.
'''
# <snippet>
import numpy as np
import GooseEYE
# generate image
I = np.zeros((21, 21), dtype='bool')
I[4, 4] = True
I[18, 19] = True
I[19, 19] = True
I[20, 19] = True
I[19, 18] = True
I[19, 20] = True
# clusters
C = GooseEYE.Clusters(I).labels()
# dilate
CD = GooseEYE.dilate(C)
# </snippet>
if __name__ == '__main__':
import docopt
args = docopt.docopt(__doc__)
if args['--save']:
import h5py
with h5py.File('clusters_dilate_periodic.h5', 'w') as data:
data['I'] = I
data['C'] = C
data['CD'] = CD
if args['--check']:
import h5py
with h5py.File('clusters_dilate_periodic.h5', 'r') as data:
assert np.all(np.equal(data['I'][...], I))
assert np.all(np.equal(data['C'][...], C))
assert np.all(np.equal(data['CD'][...], CD))
if args['--plot']:
import matplotlib.pyplot as plt
import matplotlib as mpl
import matplotlib.cm as cm
from mpl_toolkits.axes_grid1 import make_axes_locatable
# color-scheme: modify such that the background is white
# N.B. for a transparent background -> 4th column == 1.
cmap = cm.jet(range(256))
cmap[0, :3] = 1.0
cmap = mpl.colors.ListedColormap(cmap)
try:
plt.style.use(['goose', 'goose-latex'])
except:
pass
fig, axes = plt.subplots(figsize=(18, 6), nrows=1, ncols=3)
ax = axes[0]
im = ax.imshow(I, clim=(0, 1), cmap=mpl.colors.ListedColormap(cm.gray([0, 255])))
ax.xaxis.set_ticks([0, 20])
ax.yaxis.set_ticks([0, 20])
ax.set_xlim([-0.5, 20.5])
ax.set_ylim([-0.5, 20.5])
ax.set_xlabel(r'$x$')
ax.set_ylabel(r'$y$')
ax.set_title (r'image')
div = make_axes_locatable(ax)
cax = div.append_axes("right", size="5%", pad=0.1)
cbar = plt.colorbar(im, cax=cax)
cbar.set_ticks([0, 1])
ax = axes[1]
im = ax.imshow(CD, clim=(0, np.max(C) + 1), cmap=cmap)
ax.xaxis.set_ticks([0, 20])
ax.yaxis.set_ticks([0, 20])
ax.set_xlim([-0.5, 20.5])
ax.set_ylim([-0.5, 20.5])
ax.set_xlabel(r'$x$')
ax.set_ylabel(r'$y$')
ax.set_title (r'clusters + dilate')
ax = axes[2]
im = ax.imshow(np.tile(CD, (3, 3)), clim=(0, np.max(C) + 1), cmap=cmap)
ax.xaxis.set_ticks([0, 60])
ax.yaxis.set_ticks([0, 60])
ax.set_xlim([-0.5, 60.5])
ax.set_ylim([-0.5, 60.5])
ax.set_xlabel(r'$x$')
ax.set_ylabel(r'$y$')
ax.set_title (r'periodic copy')
plt.savefig('clusters_dilate_periodic.svg')
| gpl-3.0 |
aep124/TwitterAnalyticsTools | textonly.py | 1 | 2405 | # this is a script to retrieve and process text-only data for classification
# This process includes four main tasks
# 1) getting raw tweets
# 2) apply labels (this step can be conducted at any time)
# 2) filtering those tweets (e.g., according to CMU POS tagger)
# 3) deriving a set of features (a.k.a. word list)
# 4) write the feature vectors to an arff file
import tools4pgs
import tools4parsing
import tools4fv
import tools4labeling
import pickle
import copy
import numpy as np
import pandas as pd
# dividing into two dataframe because tweet info is fixed, but features are flexible
# tweet info data frame columns:
# NAME DATATYPE
# twtid ....... string (of digits)
# raw ......... string
# filtered .... string
# userid ...... string (of digits)
# handle ...... string
# label ....... string
# imgurl ...... string
# tweet features data frame columns
# twtid ....... string (of digits)
# feature 1 ... TF score for word 1
# feature 2 ... TF score for word 2
# :
# feature n ... TF score for word n
# label ....... string
############### (1) Get Tweets ################
# TODO: modify query handling to accomodate the column names that databases use, as well as subsets query variables
# (this is written for robbery database)
query = 'SELECT id,text,user_id FROM tweets'
condition = "WHERE text like '%bears%'"
tools4pgs.writetwtinfo(query, condition, 'twtinfo.p')
############### (2) Apply Labels ###############
labelmap = tools4labeling.getlabelmap('labelsystem')
tools4labeling.writelabels('twtinfo.p', labelmap)
################# (3) Filter ################
keepset = tools4parsing.getkeepset('POS2keep')
tools4parsing.writefiltered('twtinfo.p', keepset)
# TODO: add functionality for reply tweets (conversations) ????????
############## (4) Derive Features ##############
wordmap = tools4fv.getwordmap('twtinfo.p')
wordlist = wordmap.keys()
# specify threshold directly :
# freq_threshold = 2
# could also specify threshold by number of words (e.g., 500):
# freq_threshold = sorted(wordmap.values())[-500]
# wordlist = [w for w in wordmap.keys() if wordmap[w] >= freq_threshold]
tools4fv.writetf('twtinfo.p','twtfeatures.p', wordlist)
tools4fv.synclabels('twtinfo.p','twtfeatures.p')
############### (5) Make ARFF File ###############
#tools4fv.writearff('twtfeatures.p')
| mit |
felipemontefuscolo/bitme | get_bitmex_candles.py | 1 | 4122 | #!/usr/bin/env python
import sys
import time
import swagger_client
from swagger_client.rest import ApiException
from utils.utils import smart_open
import argparse
import pandas as pd
MAX_NUM_CANDLES_BITMEX = 500
def print_file(file_or_stdout, api_instance, bin_size, partial, symbol, reverse, start_time, end_time):
chunks = split_in_chunks(start_time, end_time, MAX_NUM_CANDLES_BITMEX, bin_size)
with smart_open(file_or_stdout) as fh:
print("time,open,high,low,close,volume", file=fh)
num_pages = len(chunks)
for i in range(num_pages):
chunk = chunks[i]
s = chunk[0]
e = chunk[1]
count = (e - s) / pd.Timedelta(bin_size)
page = api_instance.trade_get_bucketed(
bin_size=bin_size,
partial=partial,
symbol=symbol,
count=count,
start=0.0,
reverse=reverse,
start_time=s,
end_time=e)
print("from {} to {}: {} candles downloaded".format(s, e, len(page)))
# TODO: bitmex has a bug where the high is not the highest value !!!!!
for line in reversed(page):
print(','.join([line.timestamp.strftime('%Y-%m-%dT%H:%M:%S'),
str(line.open),
str(max(line.high, line.open)),
str(min(line.low, line.open)),
str(line.close),
str(line.volume)]), file=fh)
sys.stdout.write(
"progress: completed %d out of %d pages (%.2f%%) \r" %
(i + 1, num_pages, 100 * float(i + 1) / num_pages))
sys.stdout.flush()
time.sleep(1.001)
print("")
def split_in_chunks(start: pd.Timedelta, end: pd.Timedelta, chunk_size: int, bucket_size: str):
i = start
r = []
dt = chunk_size * pd.Timedelta(bucket_size)
while i <= end:
r += [(i, min(end, i + dt))]
i += dt
return r
def get_args(args=None, namespace=None):
parser = argparse.ArgumentParser(description="Get bitmex data")
parser.add_argument('-b', '--begin-time', type=pd.Timestamp, required=True, help="Example: '2018-04-01T00:00:01'")
parser.add_argument('-e', '--end-time', type=pd.Timestamp, required=True, help="Example: '2018-04-02T00:00:01'")
parser.add_argument('-s', '--symbol', type=str, default='XBTUSD',
help='Instrument symbol. Send a bare series (e.g. XBU) to get data for the nearest expiring'
'contract in that series. You can also send a timeframe, e.g. `XBU:monthly`. '
'Timeframes are `daily`, `weekly`, `monthly`, `quarterly`, and `biquarterly`. (optional)')
parser.add_argument('-z', '--bin-size', choices=('1m', '5m', '1h', '1d'), default='1m', type=str,
help='Time interval to bucket by')
parser.add_argument('-o', '--file-or-stdout', type=str, required=True, help='Output filename or "-" for stdout')
parser.add_argument('--partial', action='store_true', default=False, )
args = parser.parse_args(args, namespace)
return args
def main():
args = get_args()
# create an instance of the API class
configuration = swagger_client.Configuration()
configuration.host = 'https://www.bitmex.com/api/v1'
api_instance = swagger_client.TradeApi(swagger_client.ApiClient(configuration))
print("print to file " + (args.file_or_stdout if args.file_or_stdout is not '-' else 'std output'))
try:
print_file(file_or_stdout=args.file_or_stdout,
api_instance=api_instance,
bin_size=args.bin_size, partial=args.partial, symbol=args.symbol,
reverse=False,
start_time=args.begin_time, end_time=args.end_time)
except ApiException as e:
print("Exception when calling TradeApi->trade_get_bucketed: %s\n" % e)
return 0
if __name__ == "__main__":
sys.exit(main())
| mpl-2.0 |
ellio167/lammps | examples/SPIN/test_problems/validation_damped_precession/llg_precession.py | 9 | 1646 | #!/usr/bin/env python3
import numpy as np , pylab, tkinter
import math
import matplotlib.pyplot as plt
import mpmath as mp
mub=5.78901e-5 # Bohr magneton (eV/T)
hbar=0.658212 # Planck's constant (eV.fs/rad)
g=2.0 # Lande factor (adim)
gyro=g*mub/hbar # gyromag ratio (rad/fs/T)
alpha=0.01 # damping coefficient
pi=math.pi
Bnrm=10.0 # mag. field (T)
Bext = np.array([0.0, 0.0, 1.0])
Sn = 2.0 # spin norm (in # of muB)
S = np.array([1.0, 0.0, 0.0])
N=500000 # number of timesteps
dt=0.1 # timestep (fs)
# Rodrigues rotation formula
def rotation_matrix(axis, theta):
"""
Return the rotation matrix associated with counterclockwise
rotation about the given axis by theta radians
"""
axis = np.asarray(axis)
a = math.cos(theta / 2.0)
b, c, d = -axis * math.sin(theta / 2.0)
aa, bb, cc, dd = a * a, b * b, c * c, d * d
bc, ad, ac, ab, bd, cd = b * c, a * d, a * c, a * b, b * d, c * d
return np.array([[aa + bb - cc - dd, 2 * (bc + ad), 2 * (bd - ac)],
[2 * (bc - ad), aa + cc - bb - dd, 2 * (cd + ab)],
[2 * (bd + ac), 2 * (cd - ab), aa + dd - bb - cc]])
# calc. precession field
def calc_rot_vector(Fi,Sp):
rot = gyro*Sn*Bnrm*(Fi-alpha*np.cross(Fi,Sp))
return rot
# np.set_printoptions(precision=4)
for t in range (0,N):
wf = calc_rot_vector(Bext,S)
theta=dt*np.linalg.norm(wf)
axis=wf/np.linalg.norm(wf)
S = np.dot(rotation_matrix(axis, theta), S)
en = -hbar*gyro*Sn*Bnrm*np.dot(S,Bext)
# print res. in ps for comparison with LAMMPS
print(t*dt/1000.0,S[0],S[1],S[2],en)
| gpl-2.0 |
SanPen/GridCal | src/research/PTDF/ACPTDF_research2.py | 1 | 14022 | # This file is part of GridCal.
#
# GridCal is free software: you can redistribute it and/or modify
# it under the terms of the GNU General Public License as published by
# the Free Software Foundation, either version 3 of the License, or
# (at your option) any later version.
#
# GridCal is distributed in the hope that it will be useful,
# but WITHOUT ANY WARRANTY; without even the implied warranty of
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
# GNU General Public License for more details.
#
# You should have received a copy of the GNU General Public License
# along with GridCal. If not, see <http://www.gnu.org/licenses/>.
import numpy as np
import pandas as pd
import numba as nb
import time
from warnings import warn
import scipy.sparse as sp
from scipy.sparse import coo_matrix, csc_matrix
from scipy.sparse import hstack as hs, vstack as vs
from scipy.sparse.linalg import factorized, spsolve, inv
from matplotlib import pyplot as plt
from GridCal.Engine import *
def SysMat(Y, Ys, pq, pvpq):
"""
Computes the system Jacobian matrix in polar coordinates
Args:
Ybus: Admittance matrix
V: Array of nodal voltages
Ibus: Array of nodal current injections
pq: Array with the indices of the PQ buses
pvpq: Array with the indices of the PV and PQ buses
Returns:
The system Jacobian matrix
"""
A11 = -Ys.imag[np.ix_(pvpq, pvpq)]
A12 = Y.real[np.ix_(pvpq, pq)]
A21 = -Ys.real[np.ix_(pq, pvpq)]
A22 = -Y.imag[np.ix_(pq, pq)]
Asys = sp.vstack([sp.hstack([A11, A12]),
sp.hstack([A21, A22])], format="csc")
return Asys
def compute_acptdf(Ybus, Yseries, Yf, Yt, Cf, V, pq, pv, distribute_slack):
"""
Compute the AC-PTDF
:param Ybus: admittance matrix
:param Yf: Admittance matrix of the buses "from"
:param Yt: Admittance matrix of the buses "to"
:param Cf: Connectivity branch - bus "from"
:param V: voltages array
:param Ibus: array of currents
:param pq: array of pq node indices
:param pv: array of pv node indices
:return: AC-PTDF matrix (branches, buses)
"""
n = len(V)
pvpq = np.r_[pv, pq]
npq = len(pq)
# compute the Jacobian
J = SysMat(Ybus, Yseries, pq, pvpq)
if distribute_slack:
dP = np.ones((n, n)) * (-1 / (n - 1))
for i in range(n):
dP[i, i] = 1.0
else:
dP = np.eye(n, n)
# compose the compatible array (the Q increments are considered zero
dQ = np.zeros((npq, n))
# dQ = np.eye(n, n)[pq, :]
dS = np.r_[dP[pvpq, :], dQ]
# solve the voltage increments
dx = spsolve(J, dS)
# compute branch derivatives
If = Yf * V
E = V / np.abs(V)
Vdiag = sp.diags(V)
Vdiag_conj = sp.diags(np.conj(V))
Ediag = sp.diags(E)
Ediag_conj = sp.diags(np.conj(E))
If_diag_conj = sp.diags(np.conj(If))
Yf_conj = Yf.copy()
Yf_conj.data = np.conj(Yf_conj.data)
Yt_conj = Yt.copy()
Yt_conj.data = np.conj(Yt_conj.data)
dSf_dVa = 1j * (If_diag_conj * Cf * Vdiag - sp.diags(Cf * V) * Yf_conj * Vdiag_conj)
dSf_dVm = If_diag_conj * Cf * Ediag - sp.diags(Cf * V) * Yf_conj * Ediag_conj
# compose the final AC-PTDF
dPf_dVa = dSf_dVa.real[:, pvpq]
dPf_dVm = dSf_dVm.real[:, pq]
PTDF = sp.hstack((dPf_dVa, dPf_dVm)) * dx
return PTDF
def make_lodf(circuit: SnapshotCircuit, PTDF, correct_values=True):
"""
:param circuit:
:param PTDF: PTDF matrix in numpy array form
:return:
"""
nl = circuit.nbr
# compute the connectivity matrix
Cft = circuit.C_branch_bus_f - circuit.C_branch_bus_t
H = PTDF * Cft.T
# old code
# h = sp.diags(H.diagonal())
# LODF = H / (np.ones((nl, nl)) - h * np.ones(nl))
# divide each row of H by the vector 1 - H.diagonal
# LODF = H / (1 - H.diagonal())
# replace possible nan and inf
# LODF[LODF == -np.inf] = 0
# LODF[LODF == np.inf] = 0
# LODF = np.nan_to_num(LODF)
# this loop avoids the divisions by zero
# in those cases the LODF column should be zero
LODF = np.zeros((nl, nl))
div = 1 - H.diagonal()
for j in range(H.shape[1]):
if div[j] != 0:
LODF[:, j] = H[:, j] / div[j]
# replace the diagonal elements by -1
# old code
# LODF = LODF - sp.diags(LODF.diagonal()) - sp.eye(nl, nl), replaced by:
for i in range(nl):
LODF[i, i] = - 1.0
if correct_values:
i1, j1 = np.where(LODF > 1)
for i, j in zip(i1, j1):
LODF[i, j] = 1
i2, j2 = np.where(LODF < -1)
for i, j in zip(i2, j2):
LODF[i, j] = -1
return LODF
def get_branch_time_series(circuit: TimeCircuit, PTDF):
"""
:param grid:
:return:
"""
# option 2: call the power directly
P = circuit.Sbus.real
Pbr = np.dot(PTDF, P).T * circuit.Sbase
return Pbr
def multiple_failure_old(flows, LODF, beta, delta, alpha):
"""
:param flows: array of all the pre-contingency flows
:param LODF: Line Outage Distribution Factors Matrix
:param beta: index of the first failed line
:param delta: index of the second failed line
:param alpha: index of the line where you want to see the effects
:return: post contingency flow in the line alpha
"""
# multiple contingency matrix
M = np.ones((2, 2))
M[0, 1] = -LODF[beta, delta]
M[1, 0] = -LODF[delta, beta]
# normal flows of the lines beta and delta
F = flows[[beta, delta]]
# contingency flows after failing the ines beta and delta
Ff = np.linalg.solve(M, F)
# flow delta in the line alpha after the multiple contingency of the lines beta and delta
L = LODF[alpha, :][[beta, delta]]
dFf_alpha = np.dot(L, Ff)
return F[alpha] + dFf_alpha
def multiple_failure(flows, LODF, failed_idx):
"""
From the paper:
Multiple Element Contingency Screening
IEEE TRANSACTIONS ON POWER SYSTEMS, VOL. 26, NO. 3, AUGUST 2011
C. Matthew Davis and Thomas J. Overbye
:param flows: array of all the pre-contingency flows (the base flows)
:param LODF: Line Outage Distribution Factors Matrix
:param failed_idx: indices of the failed lines
:return: all post contingency flows
"""
# multiple contingency matrix
M = -LODF[np.ix_(failed_idx, failed_idx)]
for i in range(len(failed_idx)):
M[i, i] = 1.0
# normal flows of the failed lines indicated by failed_idx
F = flows[failed_idx]
# Affected flows after failing the lines indicated by failed_idx
Ff = np.linalg.solve(M, F)
# flow delta in the line alpha after the multiple contingency of the lines indicated by failed_idx
L = LODF[:, failed_idx]
dFf_alpha = np.dot(L, Ff)
# return the final contingency flow as the base flow plus the contingency flow delta
return flows + dFf_alpha
def get_n_minus_1_flows(circuit: MultiCircuit):
opt = PowerFlowOptions()
branches = circuit.get_branches()
m = circuit.get_branch_number()
Pmat = np.zeros((m, m)) # monitored, contingency
for c, branch in enumerate(branches):
if branch.active:
branch.active = False
pf = PowerFlowDriver(circuit, opt)
pf.run()
Pmat[:, c] = pf.results.Sbranch.real
branch.active = True
return Pmat
def check_lodf(grid: MultiCircuit):
flows_n1_nr = get_n_minus_1_flows(grid)
# assume 1 island
nc = compile_snapshot_circuit(grid)
islands = split_into_islands(nc)
circuit = islands[0]
pf_driver = PowerFlowDriver(grid, PowerFlowOptions())
pf_driver.run()
PTDF = compute_acptdf(Ybus=circuit.Ybus,
Yseries=circuit.Yseries,
Yf=circuit.Yf,
Yt=circuit.Yt,
Cf=circuit.C_branch_bus_f,
V=pf_driver.results.voltage,
pq=circuit.pq,
pv=circuit.pv,
distribute_slack=True)
LODF = make_lodf(circuit, PTDF)
Pbus = circuit.get_injections(False).real
flows_n = np.dot(PTDF, Pbus)
nl = circuit.nbr
flows_n1 = np.zeros((nl, nl))
for c in range(nl): # branch that fails (contingency)
# for m in range(nl): # branch to monitor
# flows_n1[m, c] = flows_n[m] + LODF[m, c] * flows_n[c]
flows_n1[:, c] = flows_n[:] + LODF[:, c] * flows_n[c]
return flows_n, flows_n1_nr, flows_n1
def test_ptdf(grid):
"""
Sigma-distances test
:param grid:
:return:
"""
nc = compile_snapshot_circuit(grid)
islands = split_into_islands(nc)
circuit = islands[0] # pick the first island
pf_driver = PowerFlowDriver(grid, PowerFlowOptions())
pf_driver.run()
PTDF = compute_acptdf(Ybus=circuit.Ybus,
Yseries=circuit.Yseries,
Yf=circuit.Yf,
Yt=circuit.Yt,
Cf=circuit.C_branch_bus_f,
V=pf_driver.results.voltage,
pq=circuit.pq,
pv=circuit.pv,
distribute_slack=False)
print('PTDF:')
print(PTDF)
if __name__ == '__main__':
from GridCal.Engine import FileOpen
import pandas as pd
np.set_printoptions(threshold=sys.maxsize, linewidth=200000000)
# np.set_printoptions(linewidth=2000, suppress=True)
pd.set_option('display.max_rows', 500)
pd.set_option('display.max_columns', 500)
pd.set_option('display.width', 1000)
# fname = '/home/santi/Documentos/GitHub/GridCal/Grids_and_profiles/grids/IEEE39_1W.gridcal'
# fname = '/home/santi/Documentos/GitHub/GridCal/Grids_and_profiles/grids/IEEE 14.xlsx'
# fname = '/home/santi/Documentos/GitHub/GridCal/Grids_and_profiles/grids/lynn5buspv.xlsx'
# fname = '/home/santi/Documentos/GitHub/GridCal/Grids_and_profiles/grids/IEEE 118.xlsx'
fname = '/home/santi/Documentos/GitHub/GridCal/Grids_and_profiles/grids/1354 Pegase.xlsx'
# fname = 'helm_data1.gridcal'
# fname = '/home/santi/Documentos/GitHub/GridCal/Grids_and_profiles/grids/IEEE 14 PQ only.gridcal'
# fname = 'IEEE 14 PQ only full.gridcal'
# fname = '/home/santi/Descargas/matpower-fubm-master/data/case5.m'
# fname = '/home/santi/Descargas/matpower-fubm-master/data/case30.m'
# fname = '/home/santi/Documentos/GitHub/GridCal/Grids_and_profiles/grids/PGOC_6bus.gridcal'
grid_ = FileOpen(fname).open()
test_ptdf(grid_)
name = os.path.splitext(fname.split(os.sep)[-1])[0]
method = 'ACPTDF (No Jacobian, V=Vpf)'
nc_ = compile_snapshot_circuit(grid_)
islands_ = split_into_islands(nc_)
circuit_ = islands_[0]
pf_driver_ = PowerFlowDriver(grid_, PowerFlowOptions())
pf_driver_.run()
H_ = compute_acptdf(Ybus=circuit_.Ybus,
Yseries=circuit_.Yseries,
Yf=circuit_.Yf,
Yt=circuit_.Yt,
Cf=circuit_.C_branch_bus_f,
V=pf_driver_.results.voltage,
pq=circuit_.pq,
pv=circuit_.pv,
distribute_slack=False)
LODF_ = make_lodf(circuit_, H_)
if H_.shape[0] < 50:
print('PTDF:\n', H_)
print('LODF:\n', LODF_)
flows_n_, flows_n1_nr_, flows_n1_ = check_lodf(grid_)
# in the case of the grid PGOC_6bus
flows_multiple = multiple_failure(flows=flows_n_,
LODF=LODF_,
failed_idx=[1, 5]) # failed lines 2 and 6
Pn1_nr_df = pd.DataFrame(data=flows_n1_nr_, index=nc_.branch_names, columns=nc_.branch_names)
flows_n1_df = pd.DataFrame(data=flows_n1_, index=nc_.branch_names, columns=nc_.branch_names)
# plot N-1
fig = plt.figure(figsize=(12, 8))
title = 'N-1 with ' + method + ' (' + name + ')'
fig.suptitle(title)
ax1 = fig.add_subplot(221)
ax2 = fig.add_subplot(222)
ax3 = fig.add_subplot(223)
Pn1_nr_df.plot(ax=ax1, legend=False)
flows_n1_df.plot(ax=ax2, legend=False)
diff = Pn1_nr_df - flows_n1_df
diff.plot(ax=ax3, legend=False)
ax1.set_title('Newton-Raphson N-1 flows')
ax2.set_title('PTDF N-1 flows')
ax3.set_title('Difference')
fig.savefig(title + '.png')
# ------------------------------------------------------------------------------------------------------------------
# Perform real time series
# ------------------------------------------------------------------------------------------------------------------
if grid_.time_profile is not None:
grid_.ensure_profiles_exist()
nc_ts = compile_time_circuit(grid_)
islands_ts = split_time_circuit_into_islands(nc_ts)
circuit_ts = islands_ts[0]
pf_options = PowerFlowOptions()
ts_driver = TimeSeries(grid=grid_, options=pf_options)
ts_driver.run()
Pbr_nr = ts_driver.results.Sbranch.real
df_Pbr_nr = pd.DataFrame(data=Pbr_nr, columns=circuit_ts.branch_names, index=circuit_ts.time_array)
# Compute the PTDF based flows
Pbr_ptdf = get_branch_time_series(circuit=circuit_ts, PTDF=H_)
df_Pbr_ptdf = pd.DataFrame(data=Pbr_ptdf, columns=circuit_ts.branch_names, index=circuit_ts.time_array)
# plot
fig = plt.figure(figsize=(12, 8))
title = 'Flows with ' + method + ' (' + name + ')'
fig.suptitle(title)
ax1 = fig.add_subplot(221)
ax2 = fig.add_subplot(222)
ax3 = fig.add_subplot(223)
df_Pbr_nr.plot(ax=ax1, legend=False)
df_Pbr_ptdf.plot(ax=ax2, legend=False)
diff = df_Pbr_nr - df_Pbr_ptdf
diff.plot(ax=ax3, legend=False)
ax1.set_title('Newton-Raphson flows')
ax2.set_title('PTDF flows')
ax3.set_title('Difference')
fig.savefig(title + '.png')
plt.show()
| gpl-3.0 |
depet/scikit-learn | sklearn/neighbors/tests/test_dist_metrics.py | 48 | 4949 | import itertools
import numpy as np
from numpy.testing import assert_array_almost_equal
import scipy
from scipy.spatial.distance import cdist
from sklearn.neighbors.dist_metrics import DistanceMetric
from nose import SkipTest
def cmp_version(version1, version2):
version1 = tuple(map(int, version1.split('.')[:2]))
version2 = tuple(map(int, version2.split('.')[:2]))
if version1 < version2:
return -1
elif version1 > version2:
return 1
else:
return 0
class TestMetrics:
def __init__(self, n1=20, n2=25, d=4, zero_frac=0.5,
rseed=0, dtype=np.float64):
np.random.seed(rseed)
self.X1 = np.random.random((n1, d)).astype(dtype)
self.X2 = np.random.random((n2, d)).astype(dtype)
# make boolean arrays: ones and zeros
self.X1_bool = self.X1.round(0)
self.X2_bool = self.X2.round(0)
V = np.random.random((d, d))
VI = np.dot(V, V.T)
self.metrics = {'euclidean': {},
'cityblock': {},
'minkowski': dict(p=(1, 1.5, 2, 3)),
'chebyshev': {},
'seuclidean': dict(V=(np.random.random(d),)),
'wminkowski': dict(p=(1, 1.5, 3),
w=(np.random.random(d),)),
'mahalanobis': dict(VI=(VI,)),
'hamming': {},
'canberra': {},
'braycurtis': {}}
self.bool_metrics = ['matching', 'jaccard', 'dice',
'kulsinski', 'rogerstanimoto', 'russellrao',
'sokalmichener', 'sokalsneath']
def test_cdist(self):
for metric, argdict in self.metrics.items():
keys = argdict.keys()
for vals in itertools.product(*argdict.values()):
kwargs = dict(zip(keys, vals))
D_true = cdist(self.X1, self.X2, metric, **kwargs)
yield self.check_cdist, metric, kwargs, D_true
for metric in self.bool_metrics:
D_true = cdist(self.X1_bool, self.X2_bool, metric)
yield self.check_cdist_bool, metric, D_true
def check_cdist(self, metric, kwargs, D_true):
if metric == 'canberra' and cmp_version(scipy.__version__, '0.9') <= 0:
raise SkipTest("Canberra distance incorrect in scipy < 0.9")
dm = DistanceMetric.get_metric(metric, **kwargs)
D12 = dm.pairwise(self.X1, self.X2)
assert_array_almost_equal(D12, D_true)
def check_cdist_bool(self, metric, D_true):
dm = DistanceMetric.get_metric(metric)
D12 = dm.pairwise(self.X1_bool, self.X2_bool)
assert_array_almost_equal(D12, D_true)
def test_pdist(self):
for metric, argdict in self.metrics.items():
keys = argdict.keys()
for vals in itertools.product(*argdict.values()):
kwargs = dict(zip(keys, vals))
D_true = cdist(self.X1, self.X1, metric, **kwargs)
yield self.check_pdist, metric, kwargs, D_true
for metric in self.bool_metrics:
D_true = cdist(self.X1_bool, self.X1_bool, metric)
yield self.check_pdist_bool, metric, D_true
def check_pdist(self, metric, kwargs, D_true):
if metric == 'canberra' and cmp_version(scipy.__version__, '0.9') <= 0:
raise SkipTest("Canberra distance incorrect in scipy < 0.9")
dm = DistanceMetric.get_metric(metric, **kwargs)
D12 = dm.pairwise(self.X1)
assert_array_almost_equal(D12, D_true)
def check_pdist_bool(self, metric, D_true):
dm = DistanceMetric.get_metric(metric)
D12 = dm.pairwise(self.X1_bool)
assert_array_almost_equal(D12, D_true)
def test_haversine_metric():
def haversine_slow(x1, x2):
return 2 * np.arcsin(np.sqrt(np.sin(0.5 * (x1[0] - x2[0])) ** 2
+ np.cos(x1[0]) * np.cos(x2[0]) *
np.sin(0.5 * (x1[1] - x2[1])) ** 2))
X = np.random.random((10, 2))
haversine = DistanceMetric.get_metric("haversine")
D1 = haversine.pairwise(X)
D2 = np.zeros_like(D1)
for i, x1 in enumerate(X):
for j, x2 in enumerate(X):
D2[i, j] = haversine_slow(x1, x2)
assert_array_almost_equal(D1, D2)
assert_array_almost_equal(haversine.dist_to_rdist(D1),
np.sin(0.5 * D2) ** 2)
def test_pyfunc_metric():
def dist_func(x1, x2, p):
return np.sum((x1 - x2) ** p) ** (1. / p)
X = np.random.random((10, 3))
euclidean = DistanceMetric.get_metric("euclidean")
pyfunc = DistanceMetric.get_metric("pyfunc", func=dist_func, p=2)
D1 = euclidean.pairwise(X)
D2 = pyfunc.pairwise(X)
assert_array_almost_equal(D1, D2)
if __name__ == '__main__':
import nose
nose.runmodule()
| bsd-3-clause |
nmartensen/pandas | pandas/tests/indexing/test_callable.py | 14 | 8721 | # -*- coding: utf-8 -*-
# pylint: disable-msg=W0612,E1101
import numpy as np
import pandas as pd
import pandas.util.testing as tm
class TestIndexingCallable(object):
def test_frame_loc_ix_callable(self):
# GH 11485
df = pd.DataFrame({'A': [1, 2, 3, 4], 'B': list('aabb'),
'C': [1, 2, 3, 4]})
# iloc cannot use boolean Series (see GH3635)
# return bool indexer
res = df.loc[lambda x: x.A > 2]
tm.assert_frame_equal(res, df.loc[df.A > 2])
res = df.loc[lambda x: x.A > 2]
tm.assert_frame_equal(res, df.loc[df.A > 2])
res = df.loc[lambda x: x.A > 2, ]
tm.assert_frame_equal(res, df.loc[df.A > 2, ])
res = df.loc[lambda x: x.A > 2, ]
tm.assert_frame_equal(res, df.loc[df.A > 2, ])
res = df.loc[lambda x: x.B == 'b', :]
tm.assert_frame_equal(res, df.loc[df.B == 'b', :])
res = df.loc[lambda x: x.B == 'b', :]
tm.assert_frame_equal(res, df.loc[df.B == 'b', :])
res = df.loc[lambda x: x.A > 2, lambda x: x.columns == 'B']
tm.assert_frame_equal(res, df.loc[df.A > 2, [False, True, False]])
res = df.loc[lambda x: x.A > 2, lambda x: x.columns == 'B']
tm.assert_frame_equal(res, df.loc[df.A > 2, [False, True, False]])
res = df.loc[lambda x: x.A > 2, lambda x: 'B']
tm.assert_series_equal(res, df.loc[df.A > 2, 'B'])
res = df.loc[lambda x: x.A > 2, lambda x: 'B']
tm.assert_series_equal(res, df.loc[df.A > 2, 'B'])
res = df.loc[lambda x: x.A > 2, lambda x: ['A', 'B']]
tm.assert_frame_equal(res, df.loc[df.A > 2, ['A', 'B']])
res = df.loc[lambda x: x.A > 2, lambda x: ['A', 'B']]
tm.assert_frame_equal(res, df.loc[df.A > 2, ['A', 'B']])
res = df.loc[lambda x: x.A == 2, lambda x: ['A', 'B']]
tm.assert_frame_equal(res, df.loc[df.A == 2, ['A', 'B']])
res = df.loc[lambda x: x.A == 2, lambda x: ['A', 'B']]
tm.assert_frame_equal(res, df.loc[df.A == 2, ['A', 'B']])
# scalar
res = df.loc[lambda x: 1, lambda x: 'A']
assert res == df.loc[1, 'A']
res = df.loc[lambda x: 1, lambda x: 'A']
assert res == df.loc[1, 'A']
def test_frame_loc_ix_callable_mixture(self):
# GH 11485
df = pd.DataFrame({'A': [1, 2, 3, 4], 'B': list('aabb'),
'C': [1, 2, 3, 4]})
res = df.loc[lambda x: x.A > 2, ['A', 'B']]
tm.assert_frame_equal(res, df.loc[df.A > 2, ['A', 'B']])
res = df.loc[lambda x: x.A > 2, ['A', 'B']]
tm.assert_frame_equal(res, df.loc[df.A > 2, ['A', 'B']])
res = df.loc[[2, 3], lambda x: ['A', 'B']]
tm.assert_frame_equal(res, df.loc[[2, 3], ['A', 'B']])
res = df.loc[[2, 3], lambda x: ['A', 'B']]
tm.assert_frame_equal(res, df.loc[[2, 3], ['A', 'B']])
res = df.loc[3, lambda x: ['A', 'B']]
tm.assert_series_equal(res, df.loc[3, ['A', 'B']])
res = df.loc[3, lambda x: ['A', 'B']]
tm.assert_series_equal(res, df.loc[3, ['A', 'B']])
def test_frame_loc_callable(self):
# GH 11485
df = pd.DataFrame({'X': [1, 2, 3, 4],
'Y': list('aabb')},
index=list('ABCD'))
# return label
res = df.loc[lambda x: ['A', 'C']]
tm.assert_frame_equal(res, df.loc[['A', 'C']])
res = df.loc[lambda x: ['A', 'C'], ]
tm.assert_frame_equal(res, df.loc[['A', 'C'], ])
res = df.loc[lambda x: ['A', 'C'], :]
tm.assert_frame_equal(res, df.loc[['A', 'C'], :])
res = df.loc[lambda x: ['A', 'C'], lambda x: 'X']
tm.assert_series_equal(res, df.loc[['A', 'C'], 'X'])
res = df.loc[lambda x: ['A', 'C'], lambda x: ['X']]
tm.assert_frame_equal(res, df.loc[['A', 'C'], ['X']])
# mixture
res = df.loc[['A', 'C'], lambda x: 'X']
tm.assert_series_equal(res, df.loc[['A', 'C'], 'X'])
res = df.loc[['A', 'C'], lambda x: ['X']]
tm.assert_frame_equal(res, df.loc[['A', 'C'], ['X']])
res = df.loc[lambda x: ['A', 'C'], 'X']
tm.assert_series_equal(res, df.loc[['A', 'C'], 'X'])
res = df.loc[lambda x: ['A', 'C'], ['X']]
tm.assert_frame_equal(res, df.loc[['A', 'C'], ['X']])
def test_frame_loc_callable_setitem(self):
# GH 11485
df = pd.DataFrame({'X': [1, 2, 3, 4],
'Y': list('aabb')},
index=list('ABCD'))
# return label
res = df.copy()
res.loc[lambda x: ['A', 'C']] = -20
exp = df.copy()
exp.loc[['A', 'C']] = -20
tm.assert_frame_equal(res, exp)
res = df.copy()
res.loc[lambda x: ['A', 'C'], :] = 20
exp = df.copy()
exp.loc[['A', 'C'], :] = 20
tm.assert_frame_equal(res, exp)
res = df.copy()
res.loc[lambda x: ['A', 'C'], lambda x: 'X'] = -1
exp = df.copy()
exp.loc[['A', 'C'], 'X'] = -1
tm.assert_frame_equal(res, exp)
res = df.copy()
res.loc[lambda x: ['A', 'C'], lambda x: ['X']] = [5, 10]
exp = df.copy()
exp.loc[['A', 'C'], ['X']] = [5, 10]
tm.assert_frame_equal(res, exp)
# mixture
res = df.copy()
res.loc[['A', 'C'], lambda x: 'X'] = np.array([-1, -2])
exp = df.copy()
exp.loc[['A', 'C'], 'X'] = np.array([-1, -2])
tm.assert_frame_equal(res, exp)
res = df.copy()
res.loc[['A', 'C'], lambda x: ['X']] = 10
exp = df.copy()
exp.loc[['A', 'C'], ['X']] = 10
tm.assert_frame_equal(res, exp)
res = df.copy()
res.loc[lambda x: ['A', 'C'], 'X'] = -2
exp = df.copy()
exp.loc[['A', 'C'], 'X'] = -2
tm.assert_frame_equal(res, exp)
res = df.copy()
res.loc[lambda x: ['A', 'C'], ['X']] = -4
exp = df.copy()
exp.loc[['A', 'C'], ['X']] = -4
tm.assert_frame_equal(res, exp)
def test_frame_iloc_callable(self):
# GH 11485
df = pd.DataFrame({'X': [1, 2, 3, 4],
'Y': list('aabb')},
index=list('ABCD'))
# return location
res = df.iloc[lambda x: [1, 3]]
tm.assert_frame_equal(res, df.iloc[[1, 3]])
res = df.iloc[lambda x: [1, 3], :]
tm.assert_frame_equal(res, df.iloc[[1, 3], :])
res = df.iloc[lambda x: [1, 3], lambda x: 0]
tm.assert_series_equal(res, df.iloc[[1, 3], 0])
res = df.iloc[lambda x: [1, 3], lambda x: [0]]
tm.assert_frame_equal(res, df.iloc[[1, 3], [0]])
# mixture
res = df.iloc[[1, 3], lambda x: 0]
tm.assert_series_equal(res, df.iloc[[1, 3], 0])
res = df.iloc[[1, 3], lambda x: [0]]
tm.assert_frame_equal(res, df.iloc[[1, 3], [0]])
res = df.iloc[lambda x: [1, 3], 0]
tm.assert_series_equal(res, df.iloc[[1, 3], 0])
res = df.iloc[lambda x: [1, 3], [0]]
tm.assert_frame_equal(res, df.iloc[[1, 3], [0]])
def test_frame_iloc_callable_setitem(self):
# GH 11485
df = pd.DataFrame({'X': [1, 2, 3, 4],
'Y': list('aabb')},
index=list('ABCD'))
# return location
res = df.copy()
res.iloc[lambda x: [1, 3]] = 0
exp = df.copy()
exp.iloc[[1, 3]] = 0
tm.assert_frame_equal(res, exp)
res = df.copy()
res.iloc[lambda x: [1, 3], :] = -1
exp = df.copy()
exp.iloc[[1, 3], :] = -1
tm.assert_frame_equal(res, exp)
res = df.copy()
res.iloc[lambda x: [1, 3], lambda x: 0] = 5
exp = df.copy()
exp.iloc[[1, 3], 0] = 5
tm.assert_frame_equal(res, exp)
res = df.copy()
res.iloc[lambda x: [1, 3], lambda x: [0]] = 25
exp = df.copy()
exp.iloc[[1, 3], [0]] = 25
tm.assert_frame_equal(res, exp)
# mixture
res = df.copy()
res.iloc[[1, 3], lambda x: 0] = -3
exp = df.copy()
exp.iloc[[1, 3], 0] = -3
tm.assert_frame_equal(res, exp)
res = df.copy()
res.iloc[[1, 3], lambda x: [0]] = -5
exp = df.copy()
exp.iloc[[1, 3], [0]] = -5
tm.assert_frame_equal(res, exp)
res = df.copy()
res.iloc[lambda x: [1, 3], 0] = 10
exp = df.copy()
exp.iloc[[1, 3], 0] = 10
tm.assert_frame_equal(res, exp)
res = df.copy()
res.iloc[lambda x: [1, 3], [0]] = [-5, -5]
exp = df.copy()
exp.iloc[[1, 3], [0]] = [-5, -5]
tm.assert_frame_equal(res, exp)
| bsd-3-clause |
sinhrks/scikit-learn | examples/hetero_feature_union.py | 288 | 6236 | """
=============================================
Feature Union with Heterogeneous Data Sources
=============================================
Datasets can often contain components of that require different feature
extraction and processing pipelines. This scenario might occur when:
1. Your dataset consists of heterogeneous data types (e.g. raster images and
text captions)
2. Your dataset is stored in a Pandas DataFrame and different columns
require different processing pipelines.
This example demonstrates how to use
:class:`sklearn.feature_extraction.FeatureUnion` on a dataset containing
different types of features. We use the 20-newsgroups dataset and compute
standard bag-of-words features for the subject line and body in separate
pipelines as well as ad hoc features on the body. We combine them (with
weights) using a FeatureUnion and finally train a classifier on the combined
set of features.
The choice of features is not particularly helpful, but serves to illustrate
the technique.
"""
# Author: Matt Terry <matt.terry@gmail.com>
#
# License: BSD 3 clause
from __future__ import print_function
import numpy as np
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.datasets import fetch_20newsgroups
from sklearn.datasets.twenty_newsgroups import strip_newsgroup_footer
from sklearn.datasets.twenty_newsgroups import strip_newsgroup_quoting
from sklearn.decomposition import TruncatedSVD
from sklearn.feature_extraction import DictVectorizer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics import classification_report
from sklearn.pipeline import FeatureUnion
from sklearn.pipeline import Pipeline
from sklearn.svm import SVC
class ItemSelector(BaseEstimator, TransformerMixin):
"""For data grouped by feature, select subset of data at a provided key.
The data is expected to be stored in a 2D data structure, where the first
index is over features and the second is over samples. i.e.
>> len(data[key]) == n_samples
Please note that this is the opposite convention to sklearn feature
matrixes (where the first index corresponds to sample).
ItemSelector only requires that the collection implement getitem
(data[key]). Examples include: a dict of lists, 2D numpy array, Pandas
DataFrame, numpy record array, etc.
>> data = {'a': [1, 5, 2, 5, 2, 8],
'b': [9, 4, 1, 4, 1, 3]}
>> ds = ItemSelector(key='a')
>> data['a'] == ds.transform(data)
ItemSelector is not designed to handle data grouped by sample. (e.g. a
list of dicts). If your data is structured this way, consider a
transformer along the lines of `sklearn.feature_extraction.DictVectorizer`.
Parameters
----------
key : hashable, required
The key corresponding to the desired value in a mappable.
"""
def __init__(self, key):
self.key = key
def fit(self, x, y=None):
return self
def transform(self, data_dict):
return data_dict[self.key]
class TextStats(BaseEstimator, TransformerMixin):
"""Extract features from each document for DictVectorizer"""
def fit(self, x, y=None):
return self
def transform(self, posts):
return [{'length': len(text),
'num_sentences': text.count('.')}
for text in posts]
class SubjectBodyExtractor(BaseEstimator, TransformerMixin):
"""Extract the subject & body from a usenet post in a single pass.
Takes a sequence of strings and produces a dict of sequences. Keys are
`subject` and `body`.
"""
def fit(self, x, y=None):
return self
def transform(self, posts):
features = np.recarray(shape=(len(posts),),
dtype=[('subject', object), ('body', object)])
for i, text in enumerate(posts):
headers, _, bod = text.partition('\n\n')
bod = strip_newsgroup_footer(bod)
bod = strip_newsgroup_quoting(bod)
features['body'][i] = bod
prefix = 'Subject:'
sub = ''
for line in headers.split('\n'):
if line.startswith(prefix):
sub = line[len(prefix):]
break
features['subject'][i] = sub
return features
pipeline = Pipeline([
# Extract the subject & body
('subjectbody', SubjectBodyExtractor()),
# Use FeatureUnion to combine the features from subject and body
('union', FeatureUnion(
transformer_list=[
# Pipeline for pulling features from the post's subject line
('subject', Pipeline([
('selector', ItemSelector(key='subject')),
('tfidf', TfidfVectorizer(min_df=50)),
])),
# Pipeline for standard bag-of-words model for body
('body_bow', Pipeline([
('selector', ItemSelector(key='body')),
('tfidf', TfidfVectorizer()),
('best', TruncatedSVD(n_components=50)),
])),
# Pipeline for pulling ad hoc features from post's body
('body_stats', Pipeline([
('selector', ItemSelector(key='body')),
('stats', TextStats()), # returns a list of dicts
('vect', DictVectorizer()), # list of dicts -> feature matrix
])),
],
# weight components in FeatureUnion
transformer_weights={
'subject': 0.8,
'body_bow': 0.5,
'body_stats': 1.0,
},
)),
# Use a SVC classifier on the combined features
('svc', SVC(kernel='linear')),
])
# limit the list of categories to make running this exmaple faster.
categories = ['alt.atheism', 'talk.religion.misc']
train = fetch_20newsgroups(random_state=1,
subset='train',
categories=categories,
)
test = fetch_20newsgroups(random_state=1,
subset='test',
categories=categories,
)
pipeline.fit(train.data, train.target)
y = pipeline.predict(test.data)
print(classification_report(y, test.target))
| bsd-3-clause |
mbayon/TFG-MachineLearning | vbig/lib/python2.7/site-packages/sklearn/model_selection/_validation.py | 4 | 53401 | """
The :mod:`sklearn.model_selection._validation` module includes classes and
functions to validate the model.
"""
# Author: Alexandre Gramfort <alexandre.gramfort@inria.fr>
# Gael Varoquaux <gael.varoquaux@normalesup.org>
# Olivier Grisel <olivier.grisel@ensta.org>
# Raghav RV <rvraghav93@gmail.com>
# License: BSD 3 clause
from __future__ import print_function
from __future__ import division
import warnings
import numbers
import time
import numpy as np
import scipy.sparse as sp
from ..base import is_classifier, clone
from ..utils import indexable, check_random_state, safe_indexing
from ..utils.deprecation import DeprecationDict
from ..utils.validation import _is_arraylike, _num_samples
from ..utils.metaestimators import _safe_split
from ..externals.joblib import Parallel, delayed, logger
from ..externals.six.moves import zip
from ..metrics.scorer import check_scoring, _check_multimetric_scoring
from ..exceptions import FitFailedWarning
from ._split import check_cv
from ..preprocessing import LabelEncoder
__all__ = ['cross_validate', 'cross_val_score', 'cross_val_predict',
'permutation_test_score', 'learning_curve', 'validation_curve']
def cross_validate(estimator, X, y=None, groups=None, scoring=None, cv=None,
n_jobs=1, verbose=0, fit_params=None,
pre_dispatch='2*n_jobs', return_train_score="warn"):
"""Evaluate metric(s) by cross-validation and also record fit/score times.
Read more in the :ref:`User Guide <multimetric_cross_validation>`.
Parameters
----------
estimator : estimator object implementing 'fit'
The object to use to fit the data.
X : array-like
The data to fit. Can be for example a list, or an array.
y : array-like, optional, default: None
The target variable to try to predict in the case of
supervised learning.
groups : array-like, with shape (n_samples,), optional
Group labels for the samples used while splitting the dataset into
train/test set.
scoring : string, callable, list/tuple, dict or None, default: None
A single string (see :ref:`scoring_parameter`) or a callable
(see :ref:`scoring`) to evaluate the predictions on the test set.
For evaluating multiple metrics, either give a list of (unique) strings
or a dict with names as keys and callables as values.
NOTE that when using custom scorers, each scorer should return a single
value. Metric functions returning a list/array of values can be wrapped
into multiple scorers that return one value each.
See :ref:`multimetric_grid_search` for an example.
If None, the estimator's default scorer (if available) is used.
cv : int, cross-validation generator or an iterable, optional
Determines the cross-validation splitting strategy.
Possible inputs for cv are:
- None, to use the default 3-fold cross validation,
- integer, to specify the number of folds in a `(Stratified)KFold`,
- An object to be used as a cross-validation generator.
- An iterable yielding train, test splits.
For integer/None inputs, if the estimator is a classifier and ``y`` is
either binary or multiclass, :class:`StratifiedKFold` is used. In all
other cases, :class:`KFold` is used.
Refer :ref:`User Guide <cross_validation>` for the various
cross-validation strategies that can be used here.
n_jobs : integer, optional
The number of CPUs to use to do the computation. -1 means
'all CPUs'.
verbose : integer, optional
The verbosity level.
fit_params : dict, optional
Parameters to pass to the fit method of the estimator.
pre_dispatch : int, or string, optional
Controls the number of jobs that get dispatched during parallel
execution. Reducing this number can be useful to avoid an
explosion of memory consumption when more jobs get dispatched
than CPUs can process. This parameter can be:
- None, in which case all the jobs are immediately
created and spawned. Use this for lightweight and
fast-running jobs, to avoid delays due to on-demand
spawning of the jobs
- An int, giving the exact number of total jobs that are
spawned
- A string, giving an expression as a function of n_jobs,
as in '2*n_jobs'
return_train_score : boolean, optional
Whether to include train scores.
Current default is ``'warn'``, which behaves as ``True`` in addition
to raising a warning when a training score is looked up.
That default will be changed to ``False`` in 0.21.
Computing training scores is used to get insights on how different
parameter settings impact the overfitting/underfitting trade-off.
However computing the scores on the training set can be computationally
expensive and is not strictly required to select the parameters that
yield the best generalization performance.
Returns
-------
scores : dict of float arrays of shape=(n_splits,)
Array of scores of the estimator for each run of the cross validation.
A dict of arrays containing the score/time arrays for each scorer is
returned. The possible keys for this ``dict`` are:
``test_score``
The score array for test scores on each cv split.
``train_score``
The score array for train scores on each cv split.
This is available only if ``return_train_score`` parameter
is ``True``.
``fit_time``
The time for fitting the estimator on the train
set for each cv split.
``score_time``
The time for scoring the estimator on the test set for each
cv split. (Note time for scoring on the train set is not
included even if ``return_train_score`` is set to ``True``
Examples
--------
>>> from sklearn import datasets, linear_model
>>> from sklearn.model_selection import cross_validate
>>> from sklearn.metrics.scorer import make_scorer
>>> from sklearn.metrics import confusion_matrix
>>> from sklearn.svm import LinearSVC
>>> diabetes = datasets.load_diabetes()
>>> X = diabetes.data[:150]
>>> y = diabetes.target[:150]
>>> lasso = linear_model.Lasso()
Single metric evaluation using ``cross_validate``
>>> cv_results = cross_validate(lasso, X, y, return_train_score=False)
>>> sorted(cv_results.keys()) # doctest: +ELLIPSIS
['fit_time', 'score_time', 'test_score']
>>> cv_results['test_score'] # doctest: +ELLIPSIS +NORMALIZE_WHITESPACE
array([ 0.33..., 0.08..., 0.03...])
Multiple metric evaluation using ``cross_validate``
(please refer the ``scoring`` parameter doc for more information)
>>> scores = cross_validate(lasso, X, y,
... scoring=('r2', 'neg_mean_squared_error'))
>>> print(scores['test_neg_mean_squared_error']) # doctest: +ELLIPSIS
[-3635.5... -3573.3... -6114.7...]
>>> print(scores['train_r2']) # doctest: +ELLIPSIS
[ 0.28... 0.39... 0.22...]
See Also
---------
:func:`sklearn.model_selection.cross_val_score`:
Run cross-validation for single metric evaluation.
:func:`sklearn.metrics.make_scorer`:
Make a scorer from a performance metric or loss function.
"""
X, y, groups = indexable(X, y, groups)
cv = check_cv(cv, y, classifier=is_classifier(estimator))
scorers, _ = _check_multimetric_scoring(estimator, scoring=scoring)
# We clone the estimator to make sure that all the folds are
# independent, and that it is pickle-able.
parallel = Parallel(n_jobs=n_jobs, verbose=verbose,
pre_dispatch=pre_dispatch)
scores = parallel(
delayed(_fit_and_score)(
clone(estimator), X, y, scorers, train, test, verbose, None,
fit_params, return_train_score=return_train_score,
return_times=True)
for train, test in cv.split(X, y, groups))
if return_train_score:
train_scores, test_scores, fit_times, score_times = zip(*scores)
train_scores = _aggregate_score_dicts(train_scores)
else:
test_scores, fit_times, score_times = zip(*scores)
test_scores = _aggregate_score_dicts(test_scores)
# TODO: replace by a dict in 0.21
ret = DeprecationDict() if return_train_score == 'warn' else {}
ret['fit_time'] = np.array(fit_times)
ret['score_time'] = np.array(score_times)
for name in scorers:
ret['test_%s' % name] = np.array(test_scores[name])
if return_train_score:
key = 'train_%s' % name
ret[key] = np.array(train_scores[name])
if return_train_score == 'warn':
message = (
'You are accessing a training score ({!r}), '
'which will not be available by default '
'any more in 0.21. If you need training scores, '
'please set return_train_score=True').format(key)
# warn on key access
ret.add_warning(key, message, FutureWarning)
return ret
def cross_val_score(estimator, X, y=None, groups=None, scoring=None, cv=None,
n_jobs=1, verbose=0, fit_params=None,
pre_dispatch='2*n_jobs'):
"""Evaluate a score by cross-validation
Read more in the :ref:`User Guide <cross_validation>`.
Parameters
----------
estimator : estimator object implementing 'fit'
The object to use to fit the data.
X : array-like
The data to fit. Can be for example a list, or an array.
y : array-like, optional, default: None
The target variable to try to predict in the case of
supervised learning.
groups : array-like, with shape (n_samples,), optional
Group labels for the samples used while splitting the dataset into
train/test set.
scoring : string, callable or None, optional, default: None
A string (see model evaluation documentation) or
a scorer callable object / function with signature
``scorer(estimator, X, y)``.
cv : int, cross-validation generator or an iterable, optional
Determines the cross-validation splitting strategy.
Possible inputs for cv are:
- None, to use the default 3-fold cross validation,
- integer, to specify the number of folds in a `(Stratified)KFold`,
- An object to be used as a cross-validation generator.
- An iterable yielding train, test splits.
For integer/None inputs, if the estimator is a classifier and ``y`` is
either binary or multiclass, :class:`StratifiedKFold` is used. In all
other cases, :class:`KFold` is used.
Refer :ref:`User Guide <cross_validation>` for the various
cross-validation strategies that can be used here.
n_jobs : integer, optional
The number of CPUs to use to do the computation. -1 means
'all CPUs'.
verbose : integer, optional
The verbosity level.
fit_params : dict, optional
Parameters to pass to the fit method of the estimator.
pre_dispatch : int, or string, optional
Controls the number of jobs that get dispatched during parallel
execution. Reducing this number can be useful to avoid an
explosion of memory consumption when more jobs get dispatched
than CPUs can process. This parameter can be:
- None, in which case all the jobs are immediately
created and spawned. Use this for lightweight and
fast-running jobs, to avoid delays due to on-demand
spawning of the jobs
- An int, giving the exact number of total jobs that are
spawned
- A string, giving an expression as a function of n_jobs,
as in '2*n_jobs'
Returns
-------
scores : array of float, shape=(len(list(cv)),)
Array of scores of the estimator for each run of the cross validation.
Examples
--------
>>> from sklearn import datasets, linear_model
>>> from sklearn.model_selection import cross_val_score
>>> diabetes = datasets.load_diabetes()
>>> X = diabetes.data[:150]
>>> y = diabetes.target[:150]
>>> lasso = linear_model.Lasso()
>>> print(cross_val_score(lasso, X, y)) # doctest: +ELLIPSIS
[ 0.33150734 0.08022311 0.03531764]
See Also
---------
:func:`sklearn.model_selection.cross_validate`:
To run cross-validation on multiple metrics and also to return
train scores, fit times and score times.
:func:`sklearn.metrics.make_scorer`:
Make a scorer from a performance metric or loss function.
"""
# To ensure multimetric format is not supported
scorer = check_scoring(estimator, scoring=scoring)
cv_results = cross_validate(estimator=estimator, X=X, y=y, groups=groups,
scoring={'score': scorer}, cv=cv,
return_train_score=False,
n_jobs=n_jobs, verbose=verbose,
fit_params=fit_params,
pre_dispatch=pre_dispatch)
return cv_results['test_score']
def _fit_and_score(estimator, X, y, scorer, train, test, verbose,
parameters, fit_params, return_train_score=False,
return_parameters=False, return_n_test_samples=False,
return_times=False, error_score='raise'):
"""Fit estimator and compute scores for a given dataset split.
Parameters
----------
estimator : estimator object implementing 'fit'
The object to use to fit the data.
X : array-like of shape at least 2D
The data to fit.
y : array-like, optional, default: None
The target variable to try to predict in the case of
supervised learning.
scorer : A single callable or dict mapping scorer name to the callable
If it is a single callable, the return value for ``train_scores`` and
``test_scores`` is a single float.
For a dict, it should be one mapping the scorer name to the scorer
callable object / function.
The callable object / fn should have signature
``scorer(estimator, X, y)``.
train : array-like, shape (n_train_samples,)
Indices of training samples.
test : array-like, shape (n_test_samples,)
Indices of test samples.
verbose : integer
The verbosity level.
error_score : 'raise' (default) or numeric
Value to assign to the score if an error occurs in estimator fitting.
If set to 'raise', the error is raised. If a numeric value is given,
FitFailedWarning is raised. This parameter does not affect the refit
step, which will always raise the error.
parameters : dict or None
Parameters to be set on the estimator.
fit_params : dict or None
Parameters that will be passed to ``estimator.fit``.
return_train_score : boolean, optional, default: False
Compute and return score on training set.
return_parameters : boolean, optional, default: False
Return parameters that has been used for the estimator.
return_n_test_samples : boolean, optional, default: False
Whether to return the ``n_test_samples``
return_times : boolean, optional, default: False
Whether to return the fit/score times.
Returns
-------
train_scores : dict of scorer name -> float, optional
Score on training set (for all the scorers),
returned only if `return_train_score` is `True`.
test_scores : dict of scorer name -> float, optional
Score on testing set (for all the scorers).
n_test_samples : int
Number of test samples.
fit_time : float
Time spent for fitting in seconds.
score_time : float
Time spent for scoring in seconds.
parameters : dict or None, optional
The parameters that have been evaluated.
"""
if verbose > 1:
if parameters is None:
msg = ''
else:
msg = '%s' % (', '.join('%s=%s' % (k, v)
for k, v in parameters.items()))
print("[CV] %s %s" % (msg, (64 - len(msg)) * '.'))
# Adjust length of sample weights
fit_params = fit_params if fit_params is not None else {}
fit_params = dict([(k, _index_param_value(X, v, train))
for k, v in fit_params.items()])
test_scores = {}
train_scores = {}
if parameters is not None:
estimator.set_params(**parameters)
start_time = time.time()
X_train, y_train = _safe_split(estimator, X, y, train)
X_test, y_test = _safe_split(estimator, X, y, test, train)
is_multimetric = not callable(scorer)
n_scorers = len(scorer.keys()) if is_multimetric else 1
try:
if y_train is None:
estimator.fit(X_train, **fit_params)
else:
estimator.fit(X_train, y_train, **fit_params)
except Exception as e:
# Note fit time as time until error
fit_time = time.time() - start_time
score_time = 0.0
if error_score == 'raise':
raise
elif isinstance(error_score, numbers.Number):
if is_multimetric:
test_scores = dict(zip(scorer.keys(),
[error_score, ] * n_scorers))
if return_train_score:
train_scores = dict(zip(scorer.keys(),
[error_score, ] * n_scorers))
else:
test_scores = error_score
if return_train_score:
train_scores = error_score
warnings.warn("Classifier fit failed. The score on this train-test"
" partition for these parameters will be set to %f. "
"Details: \n%r" % (error_score, e), FitFailedWarning)
else:
raise ValueError("error_score must be the string 'raise' or a"
" numeric value. (Hint: if using 'raise', please"
" make sure that it has been spelled correctly.)")
else:
fit_time = time.time() - start_time
# _score will return dict if is_multimetric is True
test_scores = _score(estimator, X_test, y_test, scorer, is_multimetric)
score_time = time.time() - start_time - fit_time
if return_train_score:
train_scores = _score(estimator, X_train, y_train, scorer,
is_multimetric)
if verbose > 2:
if is_multimetric:
for scorer_name, score in test_scores.items():
msg += ", %s=%s" % (scorer_name, score)
else:
msg += ", score=%s" % test_scores
if verbose > 1:
total_time = score_time + fit_time
end_msg = "%s, total=%s" % (msg, logger.short_format_time(total_time))
print("[CV] %s %s" % ((64 - len(end_msg)) * '.', end_msg))
ret = [train_scores, test_scores] if return_train_score else [test_scores]
if return_n_test_samples:
ret.append(_num_samples(X_test))
if return_times:
ret.extend([fit_time, score_time])
if return_parameters:
ret.append(parameters)
return ret
def _score(estimator, X_test, y_test, scorer, is_multimetric=False):
"""Compute the score(s) of an estimator on a given test set.
Will return a single float if is_multimetric is False and a dict of floats,
if is_multimetric is True
"""
if is_multimetric:
return _multimetric_score(estimator, X_test, y_test, scorer)
else:
if y_test is None:
score = scorer(estimator, X_test)
else:
score = scorer(estimator, X_test, y_test)
if hasattr(score, 'item'):
try:
# e.g. unwrap memmapped scalars
score = score.item()
except ValueError:
# non-scalar?
pass
if not isinstance(score, numbers.Number):
raise ValueError("scoring must return a number, got %s (%s) "
"instead. (scorer=%r)"
% (str(score), type(score), scorer))
return score
def _multimetric_score(estimator, X_test, y_test, scorers):
"""Return a dict of score for multimetric scoring"""
scores = {}
for name, scorer in scorers.items():
if y_test is None:
score = scorer(estimator, X_test)
else:
score = scorer(estimator, X_test, y_test)
if hasattr(score, 'item'):
try:
# e.g. unwrap memmapped scalars
score = score.item()
except ValueError:
# non-scalar?
pass
scores[name] = score
if not isinstance(score, numbers.Number):
raise ValueError("scoring must return a number, got %s (%s) "
"instead. (scorer=%s)"
% (str(score), type(score), name))
return scores
def cross_val_predict(estimator, X, y=None, groups=None, cv=None, n_jobs=1,
verbose=0, fit_params=None, pre_dispatch='2*n_jobs',
method='predict'):
"""Generate cross-validated estimates for each input data point
Read more in the :ref:`User Guide <cross_validation>`.
Parameters
----------
estimator : estimator object implementing 'fit' and 'predict'
The object to use to fit the data.
X : array-like
The data to fit. Can be, for example a list, or an array at least 2d.
y : array-like, optional, default: None
The target variable to try to predict in the case of
supervised learning.
groups : array-like, with shape (n_samples,), optional
Group labels for the samples used while splitting the dataset into
train/test set.
cv : int, cross-validation generator or an iterable, optional
Determines the cross-validation splitting strategy.
Possible inputs for cv are:
- None, to use the default 3-fold cross validation,
- integer, to specify the number of folds in a `(Stratified)KFold`,
- An object to be used as a cross-validation generator.
- An iterable yielding train, test splits.
For integer/None inputs, if the estimator is a classifier and ``y`` is
either binary or multiclass, :class:`StratifiedKFold` is used. In all
other cases, :class:`KFold` is used.
Refer :ref:`User Guide <cross_validation>` for the various
cross-validation strategies that can be used here.
n_jobs : integer, optional
The number of CPUs to use to do the computation. -1 means
'all CPUs'.
verbose : integer, optional
The verbosity level.
fit_params : dict, optional
Parameters to pass to the fit method of the estimator.
pre_dispatch : int, or string, optional
Controls the number of jobs that get dispatched during parallel
execution. Reducing this number can be useful to avoid an
explosion of memory consumption when more jobs get dispatched
than CPUs can process. This parameter can be:
- None, in which case all the jobs are immediately
created and spawned. Use this for lightweight and
fast-running jobs, to avoid delays due to on-demand
spawning of the jobs
- An int, giving the exact number of total jobs that are
spawned
- A string, giving an expression as a function of n_jobs,
as in '2*n_jobs'
method : string, optional, default: 'predict'
Invokes the passed method name of the passed estimator. For
method='predict_proba', the columns correspond to the classes
in sorted order.
Returns
-------
predictions : ndarray
This is the result of calling ``method``
Notes
-----
In the case that one or more classes are absent in a training portion, a
default score needs to be assigned to all instances for that class if
``method`` produces columns per class, as in {'decision_function',
'predict_proba', 'predict_log_proba'}. For ``predict_proba`` this value is
0. In order to ensure finite output, we approximate negative infinity by
the minimum finite float value for the dtype in other cases.
Examples
--------
>>> from sklearn import datasets, linear_model
>>> from sklearn.model_selection import cross_val_predict
>>> diabetes = datasets.load_diabetes()
>>> X = diabetes.data[:150]
>>> y = diabetes.target[:150]
>>> lasso = linear_model.Lasso()
>>> y_pred = cross_val_predict(lasso, X, y)
"""
X, y, groups = indexable(X, y, groups)
cv = check_cv(cv, y, classifier=is_classifier(estimator))
if method in ['decision_function', 'predict_proba', 'predict_log_proba']:
le = LabelEncoder()
y = le.fit_transform(y)
# We clone the estimator to make sure that all the folds are
# independent, and that it is pickle-able.
parallel = Parallel(n_jobs=n_jobs, verbose=verbose,
pre_dispatch=pre_dispatch)
prediction_blocks = parallel(delayed(_fit_and_predict)(
clone(estimator), X, y, train, test, verbose, fit_params, method)
for train, test in cv.split(X, y, groups))
# Concatenate the predictions
predictions = [pred_block_i for pred_block_i, _ in prediction_blocks]
test_indices = np.concatenate([indices_i
for _, indices_i in prediction_blocks])
if not _check_is_permutation(test_indices, _num_samples(X)):
raise ValueError('cross_val_predict only works for partitions')
inv_test_indices = np.empty(len(test_indices), dtype=int)
inv_test_indices[test_indices] = np.arange(len(test_indices))
# Check for sparse predictions
if sp.issparse(predictions[0]):
predictions = sp.vstack(predictions, format=predictions[0].format)
else:
predictions = np.concatenate(predictions)
return predictions[inv_test_indices]
def _fit_and_predict(estimator, X, y, train, test, verbose, fit_params,
method):
"""Fit estimator and predict values for a given dataset split.
Read more in the :ref:`User Guide <cross_validation>`.
Parameters
----------
estimator : estimator object implementing 'fit' and 'predict'
The object to use to fit the data.
X : array-like of shape at least 2D
The data to fit.
y : array-like, optional, default: None
The target variable to try to predict in the case of
supervised learning.
train : array-like, shape (n_train_samples,)
Indices of training samples.
test : array-like, shape (n_test_samples,)
Indices of test samples.
verbose : integer
The verbosity level.
fit_params : dict or None
Parameters that will be passed to ``estimator.fit``.
method : string
Invokes the passed method name of the passed estimator.
Returns
-------
predictions : sequence
Result of calling 'estimator.method'
test : array-like
This is the value of the test parameter
"""
# Adjust length of sample weights
fit_params = fit_params if fit_params is not None else {}
fit_params = dict([(k, _index_param_value(X, v, train))
for k, v in fit_params.items()])
X_train, y_train = _safe_split(estimator, X, y, train)
X_test, _ = _safe_split(estimator, X, y, test, train)
if y_train is None:
estimator.fit(X_train, **fit_params)
else:
estimator.fit(X_train, y_train, **fit_params)
func = getattr(estimator, method)
predictions = func(X_test)
if method in ['decision_function', 'predict_proba', 'predict_log_proba']:
n_classes = len(set(y))
if n_classes != len(estimator.classes_):
recommendation = (
'To fix this, use a cross-validation '
'technique resulting in properly '
'stratified folds')
warnings.warn('Number of classes in training fold ({}) does '
'not match total number of classes ({}). '
'Results may not be appropriate for your use case. '
'{}'.format(len(estimator.classes_),
n_classes, recommendation),
RuntimeWarning)
if method == 'decision_function':
if (predictions.ndim == 2 and
predictions.shape[1] != len(estimator.classes_)):
# This handles the case when the shape of predictions
# does not match the number of classes used to train
# it with. This case is found when sklearn.svm.SVC is
# set to `decision_function_shape='ovo'`.
raise ValueError('Output shape {} of {} does not match '
'number of classes ({}) in fold. '
'Irregular decision_function outputs '
'are not currently supported by '
'cross_val_predict'.format(
predictions.shape, method,
len(estimator.classes_),
recommendation))
if len(estimator.classes_) <= 2:
# In this special case, `predictions` contains a 1D array.
raise ValueError('Only {} class/es in training fold, this '
'is not supported for decision_function '
'with imbalanced folds. {}'.format(
len(estimator.classes_),
recommendation))
float_min = np.finfo(predictions.dtype).min
default_values = {'decision_function': float_min,
'predict_log_proba': float_min,
'predict_proba': 0}
predictions_for_all_classes = np.full((_num_samples(predictions),
n_classes),
default_values[method])
predictions_for_all_classes[:, estimator.classes_] = predictions
predictions = predictions_for_all_classes
return predictions, test
def _check_is_permutation(indices, n_samples):
"""Check whether indices is a reordering of the array np.arange(n_samples)
Parameters
----------
indices : ndarray
integer array to test
n_samples : int
number of expected elements
Returns
-------
is_partition : bool
True iff sorted(indices) is np.arange(n)
"""
if len(indices) != n_samples:
return False
hit = np.zeros(n_samples, dtype=bool)
hit[indices] = True
if not np.all(hit):
return False
return True
def _index_param_value(X, v, indices):
"""Private helper function for parameter value indexing."""
if not _is_arraylike(v) or _num_samples(v) != _num_samples(X):
# pass through: skip indexing
return v
if sp.issparse(v):
v = v.tocsr()
return safe_indexing(v, indices)
def permutation_test_score(estimator, X, y, groups=None, cv=None,
n_permutations=100, n_jobs=1, random_state=0,
verbose=0, scoring=None):
"""Evaluate the significance of a cross-validated score with permutations
Read more in the :ref:`User Guide <cross_validation>`.
Parameters
----------
estimator : estimator object implementing 'fit'
The object to use to fit the data.
X : array-like of shape at least 2D
The data to fit.
y : array-like
The target variable to try to predict in the case of
supervised learning.
groups : array-like, with shape (n_samples,), optional
Labels to constrain permutation within groups, i.e. ``y`` values
are permuted among samples with the same group identifier.
When not specified, ``y`` values are permuted among all samples.
When a grouped cross-validator is used, the group labels are
also passed on to the ``split`` method of the cross-validator. The
cross-validator uses them for grouping the samples while splitting
the dataset into train/test set.
scoring : string, callable or None, optional, default: None
A single string (see :ref:`scoring_parameter`) or a callable
(see :ref:`scoring`) to evaluate the predictions on the test set.
If None the estimator's default scorer, if available, is used.
cv : int, cross-validation generator or an iterable, optional
Determines the cross-validation splitting strategy.
Possible inputs for cv are:
- None, to use the default 3-fold cross validation,
- integer, to specify the number of folds in a `(Stratified)KFold`,
- An object to be used as a cross-validation generator.
- An iterable yielding train, test splits.
For integer/None inputs, if the estimator is a classifier and ``y`` is
either binary or multiclass, :class:`StratifiedKFold` is used. In all
other cases, :class:`KFold` is used.
Refer :ref:`User Guide <cross_validation>` for the various
cross-validation strategies that can be used here.
n_permutations : integer, optional
Number of times to permute ``y``.
n_jobs : integer, optional
The number of CPUs to use to do the computation. -1 means
'all CPUs'.
random_state : int, RandomState instance or None, optional (default=0)
If int, random_state is the seed used by the random number generator;
If RandomState instance, random_state is the random number generator;
If None, the random number generator is the RandomState instance used
by `np.random`.
verbose : integer, optional
The verbosity level.
Returns
-------
score : float
The true score without permuting targets.
permutation_scores : array, shape (n_permutations,)
The scores obtained for each permutations.
pvalue : float
The p-value, which approximates the probability that the score would
be obtained by chance. This is calculated as:
`(C + 1) / (n_permutations + 1)`
Where C is the number of permutations whose score >= the true score.
The best possible p-value is 1/(n_permutations + 1), the worst is 1.0.
Notes
-----
This function implements Test 1 in:
Ojala and Garriga. Permutation Tests for Studying Classifier
Performance. The Journal of Machine Learning Research (2010)
vol. 11
"""
X, y, groups = indexable(X, y, groups)
cv = check_cv(cv, y, classifier=is_classifier(estimator))
scorer = check_scoring(estimator, scoring=scoring)
random_state = check_random_state(random_state)
# We clone the estimator to make sure that all the folds are
# independent, and that it is pickle-able.
score = _permutation_test_score(clone(estimator), X, y, groups, cv, scorer)
permutation_scores = Parallel(n_jobs=n_jobs, verbose=verbose)(
delayed(_permutation_test_score)(
clone(estimator), X, _shuffle(y, groups, random_state),
groups, cv, scorer)
for _ in range(n_permutations))
permutation_scores = np.array(permutation_scores)
pvalue = (np.sum(permutation_scores >= score) + 1.0) / (n_permutations + 1)
return score, permutation_scores, pvalue
permutation_test_score.__test__ = False # to avoid a pb with nosetests
def _permutation_test_score(estimator, X, y, groups, cv, scorer):
"""Auxiliary function for permutation_test_score"""
avg_score = []
for train, test in cv.split(X, y, groups):
X_train, y_train = _safe_split(estimator, X, y, train)
X_test, y_test = _safe_split(estimator, X, y, test, train)
estimator.fit(X_train, y_train)
avg_score.append(scorer(estimator, X_test, y_test))
return np.mean(avg_score)
def _shuffle(y, groups, random_state):
"""Return a shuffled copy of y eventually shuffle among same groups."""
if groups is None:
indices = random_state.permutation(len(y))
else:
indices = np.arange(len(groups))
for group in np.unique(groups):
this_mask = (groups == group)
indices[this_mask] = random_state.permutation(indices[this_mask])
return safe_indexing(y, indices)
def learning_curve(estimator, X, y, groups=None,
train_sizes=np.linspace(0.1, 1.0, 5), cv=None, scoring=None,
exploit_incremental_learning=False, n_jobs=1,
pre_dispatch="all", verbose=0, shuffle=False,
random_state=None):
"""Learning curve.
Determines cross-validated training and test scores for different training
set sizes.
A cross-validation generator splits the whole dataset k times in training
and test data. Subsets of the training set with varying sizes will be used
to train the estimator and a score for each training subset size and the
test set will be computed. Afterwards, the scores will be averaged over
all k runs for each training subset size.
Read more in the :ref:`User Guide <learning_curve>`.
Parameters
----------
estimator : object type that implements the "fit" and "predict" methods
An object of that type which is cloned for each validation.
X : array-like, shape (n_samples, n_features)
Training vector, where n_samples is the number of samples and
n_features is the number of features.
y : array-like, shape (n_samples) or (n_samples, n_features), optional
Target relative to X for classification or regression;
None for unsupervised learning.
groups : array-like, with shape (n_samples,), optional
Group labels for the samples used while splitting the dataset into
train/test set.
train_sizes : array-like, shape (n_ticks,), dtype float or int
Relative or absolute numbers of training examples that will be used to
generate the learning curve. If the dtype is float, it is regarded as a
fraction of the maximum size of the training set (that is determined
by the selected validation method), i.e. it has to be within (0, 1].
Otherwise it is interpreted as absolute sizes of the training sets.
Note that for classification the number of samples usually have to
be big enough to contain at least one sample from each class.
(default: np.linspace(0.1, 1.0, 5))
cv : int, cross-validation generator or an iterable, optional
Determines the cross-validation splitting strategy.
Possible inputs for cv are:
- None, to use the default 3-fold cross validation,
- integer, to specify the number of folds in a `(Stratified)KFold`,
- An object to be used as a cross-validation generator.
- An iterable yielding train, test splits.
For integer/None inputs, if the estimator is a classifier and ``y`` is
either binary or multiclass, :class:`StratifiedKFold` is used. In all
other cases, :class:`KFold` is used.
Refer :ref:`User Guide <cross_validation>` for the various
cross-validation strategies that can be used here.
scoring : string, callable or None, optional, default: None
A string (see model evaluation documentation) or
a scorer callable object / function with signature
``scorer(estimator, X, y)``.
exploit_incremental_learning : boolean, optional, default: False
If the estimator supports incremental learning, this will be
used to speed up fitting for different training set sizes.
n_jobs : integer, optional
Number of jobs to run in parallel (default 1).
pre_dispatch : integer or string, optional
Number of predispatched jobs for parallel execution (default is
all). The option can reduce the allocated memory. The string can
be an expression like '2*n_jobs'.
verbose : integer, optional
Controls the verbosity: the higher, the more messages.
shuffle : boolean, optional
Whether to shuffle training data before taking prefixes of it
based on``train_sizes``.
random_state : int, RandomState instance or None, optional (default=None)
If int, random_state is the seed used by the random number generator;
If RandomState instance, random_state is the random number generator;
If None, the random number generator is the RandomState instance used
by `np.random`. Used when ``shuffle`` is True.
Returns
-------
train_sizes_abs : array, shape = (n_unique_ticks,), dtype int
Numbers of training examples that has been used to generate the
learning curve. Note that the number of ticks might be less
than n_ticks because duplicate entries will be removed.
train_scores : array, shape (n_ticks, n_cv_folds)
Scores on training sets.
test_scores : array, shape (n_ticks, n_cv_folds)
Scores on test set.
Notes
-----
See :ref:`examples/model_selection/plot_learning_curve.py
<sphx_glr_auto_examples_model_selection_plot_learning_curve.py>`
"""
if exploit_incremental_learning and not hasattr(estimator, "partial_fit"):
raise ValueError("An estimator must support the partial_fit interface "
"to exploit incremental learning")
X, y, groups = indexable(X, y, groups)
cv = check_cv(cv, y, classifier=is_classifier(estimator))
# Store it as list as we will be iterating over the list multiple times
cv_iter = list(cv.split(X, y, groups))
scorer = check_scoring(estimator, scoring=scoring)
n_max_training_samples = len(cv_iter[0][0])
# Because the lengths of folds can be significantly different, it is
# not guaranteed that we use all of the available training data when we
# use the first 'n_max_training_samples' samples.
train_sizes_abs = _translate_train_sizes(train_sizes,
n_max_training_samples)
n_unique_ticks = train_sizes_abs.shape[0]
if verbose > 0:
print("[learning_curve] Training set sizes: " + str(train_sizes_abs))
parallel = Parallel(n_jobs=n_jobs, pre_dispatch=pre_dispatch,
verbose=verbose)
if shuffle:
rng = check_random_state(random_state)
cv_iter = ((rng.permutation(train), test) for train, test in cv_iter)
if exploit_incremental_learning:
classes = np.unique(y) if is_classifier(estimator) else None
out = parallel(delayed(_incremental_fit_estimator)(
clone(estimator), X, y, classes, train, test, train_sizes_abs,
scorer, verbose) for train, test in cv_iter)
else:
train_test_proportions = []
for train, test in cv_iter:
for n_train_samples in train_sizes_abs:
train_test_proportions.append((train[:n_train_samples], test))
out = parallel(delayed(_fit_and_score)(
clone(estimator), X, y, scorer, train, test,
verbose, parameters=None, fit_params=None, return_train_score=True)
for train, test in train_test_proportions)
out = np.array(out)
n_cv_folds = out.shape[0] // n_unique_ticks
out = out.reshape(n_cv_folds, n_unique_ticks, 2)
out = np.asarray(out).transpose((2, 1, 0))
return train_sizes_abs, out[0], out[1]
def _translate_train_sizes(train_sizes, n_max_training_samples):
"""Determine absolute sizes of training subsets and validate 'train_sizes'.
Examples:
_translate_train_sizes([0.5, 1.0], 10) -> [5, 10]
_translate_train_sizes([5, 10], 10) -> [5, 10]
Parameters
----------
train_sizes : array-like, shape (n_ticks,), dtype float or int
Numbers of training examples that will be used to generate the
learning curve. If the dtype is float, it is regarded as a
fraction of 'n_max_training_samples', i.e. it has to be within (0, 1].
n_max_training_samples : int
Maximum number of training samples (upper bound of 'train_sizes').
Returns
-------
train_sizes_abs : array, shape (n_unique_ticks,), dtype int
Numbers of training examples that will be used to generate the
learning curve. Note that the number of ticks might be less
than n_ticks because duplicate entries will be removed.
"""
train_sizes_abs = np.asarray(train_sizes)
n_ticks = train_sizes_abs.shape[0]
n_min_required_samples = np.min(train_sizes_abs)
n_max_required_samples = np.max(train_sizes_abs)
if np.issubdtype(train_sizes_abs.dtype, np.floating):
if n_min_required_samples <= 0.0 or n_max_required_samples > 1.0:
raise ValueError("train_sizes has been interpreted as fractions "
"of the maximum number of training samples and "
"must be within (0, 1], but is within [%f, %f]."
% (n_min_required_samples,
n_max_required_samples))
train_sizes_abs = (train_sizes_abs * n_max_training_samples).astype(
dtype=np.int, copy=False)
train_sizes_abs = np.clip(train_sizes_abs, 1,
n_max_training_samples)
else:
if (n_min_required_samples <= 0 or
n_max_required_samples > n_max_training_samples):
raise ValueError("train_sizes has been interpreted as absolute "
"numbers of training samples and must be within "
"(0, %d], but is within [%d, %d]."
% (n_max_training_samples,
n_min_required_samples,
n_max_required_samples))
train_sizes_abs = np.unique(train_sizes_abs)
if n_ticks > train_sizes_abs.shape[0]:
warnings.warn("Removed duplicate entries from 'train_sizes'. Number "
"of ticks will be less than the size of "
"'train_sizes' %d instead of %d)."
% (train_sizes_abs.shape[0], n_ticks), RuntimeWarning)
return train_sizes_abs
def _incremental_fit_estimator(estimator, X, y, classes, train, test,
train_sizes, scorer, verbose):
"""Train estimator on training subsets incrementally and compute scores."""
train_scores, test_scores = [], []
partitions = zip(train_sizes, np.split(train, train_sizes)[:-1])
for n_train_samples, partial_train in partitions:
train_subset = train[:n_train_samples]
X_train, y_train = _safe_split(estimator, X, y, train_subset)
X_partial_train, y_partial_train = _safe_split(estimator, X, y,
partial_train)
X_test, y_test = _safe_split(estimator, X, y, test, train_subset)
if y_partial_train is None:
estimator.partial_fit(X_partial_train, classes=classes)
else:
estimator.partial_fit(X_partial_train, y_partial_train,
classes=classes)
train_scores.append(_score(estimator, X_train, y_train, scorer))
test_scores.append(_score(estimator, X_test, y_test, scorer))
return np.array((train_scores, test_scores)).T
def validation_curve(estimator, X, y, param_name, param_range, groups=None,
cv=None, scoring=None, n_jobs=1, pre_dispatch="all",
verbose=0):
"""Validation curve.
Determine training and test scores for varying parameter values.
Compute scores for an estimator with different values of a specified
parameter. This is similar to grid search with one parameter. However, this
will also compute training scores and is merely a utility for plotting the
results.
Read more in the :ref:`User Guide <learning_curve>`.
Parameters
----------
estimator : object type that implements the "fit" and "predict" methods
An object of that type which is cloned for each validation.
X : array-like, shape (n_samples, n_features)
Training vector, where n_samples is the number of samples and
n_features is the number of features.
y : array-like, shape (n_samples) or (n_samples, n_features), optional
Target relative to X for classification or regression;
None for unsupervised learning.
param_name : string
Name of the parameter that will be varied.
param_range : array-like, shape (n_values,)
The values of the parameter that will be evaluated.
groups : array-like, with shape (n_samples,), optional
Group labels for the samples used while splitting the dataset into
train/test set.
cv : int, cross-validation generator or an iterable, optional
Determines the cross-validation splitting strategy.
Possible inputs for cv are:
- None, to use the default 3-fold cross validation,
- integer, to specify the number of folds in a `(Stratified)KFold`,
- An object to be used as a cross-validation generator.
- An iterable yielding train, test splits.
For integer/None inputs, if the estimator is a classifier and ``y`` is
either binary or multiclass, :class:`StratifiedKFold` is used. In all
other cases, :class:`KFold` is used.
Refer :ref:`User Guide <cross_validation>` for the various
cross-validation strategies that can be used here.
scoring : string, callable or None, optional, default: None
A string (see model evaluation documentation) or
a scorer callable object / function with signature
``scorer(estimator, X, y)``.
n_jobs : integer, optional
Number of jobs to run in parallel (default 1).
pre_dispatch : integer or string, optional
Number of predispatched jobs for parallel execution (default is
all). The option can reduce the allocated memory. The string can
be an expression like '2*n_jobs'.
verbose : integer, optional
Controls the verbosity: the higher, the more messages.
Returns
-------
train_scores : array, shape (n_ticks, n_cv_folds)
Scores on training sets.
test_scores : array, shape (n_ticks, n_cv_folds)
Scores on test set.
Notes
-----
See :ref:`sphx_glr_auto_examples_model_selection_plot_validation_curve.py`
"""
X, y, groups = indexable(X, y, groups)
cv = check_cv(cv, y, classifier=is_classifier(estimator))
scorer = check_scoring(estimator, scoring=scoring)
parallel = Parallel(n_jobs=n_jobs, pre_dispatch=pre_dispatch,
verbose=verbose)
out = parallel(delayed(_fit_and_score)(
clone(estimator), X, y, scorer, train, test, verbose,
parameters={param_name: v}, fit_params=None, return_train_score=True)
# NOTE do not change order of iteration to allow one time cv splitters
for train, test in cv.split(X, y, groups) for v in param_range)
out = np.asarray(out)
n_params = len(param_range)
n_cv_folds = out.shape[0] // n_params
out = out.reshape(n_cv_folds, n_params, 2).transpose((2, 1, 0))
return out[0], out[1]
def _aggregate_score_dicts(scores):
"""Aggregate the list of dict to dict of np ndarray
The aggregated output of _fit_and_score will be a list of dict
of form [{'prec': 0.1, 'acc':1.0}, {'prec': 0.1, 'acc':1.0}, ...]
Convert it to a dict of array {'prec': np.array([0.1 ...]), ...}
Parameters
----------
scores : list of dict
List of dicts of the scores for all scorers. This is a flat list,
assumed originally to be of row major order.
Example
-------
>>> scores = [{'a': 1, 'b':10}, {'a': 2, 'b':2}, {'a': 3, 'b':3},
... {'a': 10, 'b': 10}] # doctest: +SKIP
>>> _aggregate_score_dicts(scores) # doctest: +SKIP
{'a': array([1, 2, 3, 10]),
'b': array([10, 2, 3, 10])}
"""
out = {}
for key in scores[0]:
out[key] = np.asarray([score[key] for score in scores])
return out
| mit |
quheng/scikit-learn | examples/decomposition/plot_image_denoising.py | 181 | 5819 | """
=========================================
Image denoising using dictionary learning
=========================================
An example comparing the effect of reconstructing noisy fragments
of the Lena image using firstly online :ref:`DictionaryLearning` and
various transform methods.
The dictionary is fitted on the distorted left half of the image, and
subsequently used to reconstruct the right half. Note that even better
performance could be achieved by fitting to an undistorted (i.e.
noiseless) image, but here we start from the assumption that it is not
available.
A common practice for evaluating the results of image denoising is by looking
at the difference between the reconstruction and the original image. If the
reconstruction is perfect this will look like Gaussian noise.
It can be seen from the plots that the results of :ref:`omp` with two
non-zero coefficients is a bit less biased than when keeping only one
(the edges look less prominent). It is in addition closer from the ground
truth in Frobenius norm.
The result of :ref:`least_angle_regression` is much more strongly biased: the
difference is reminiscent of the local intensity value of the original image.
Thresholding is clearly not useful for denoising, but it is here to show that
it can produce a suggestive output with very high speed, and thus be useful
for other tasks such as object classification, where performance is not
necessarily related to visualisation.
"""
print(__doc__)
from time import time
import matplotlib.pyplot as plt
import numpy as np
from scipy.misc import lena
from sklearn.decomposition import MiniBatchDictionaryLearning
from sklearn.feature_extraction.image import extract_patches_2d
from sklearn.feature_extraction.image import reconstruct_from_patches_2d
###############################################################################
# Load Lena image and extract patches
lena = lena() / 256.0
# downsample for higher speed
lena = lena[::2, ::2] + lena[1::2, ::2] + lena[::2, 1::2] + lena[1::2, 1::2]
lena /= 4.0
height, width = lena.shape
# Distort the right half of the image
print('Distorting image...')
distorted = lena.copy()
distorted[:, height // 2:] += 0.075 * np.random.randn(width, height // 2)
# Extract all reference patches from the left half of the image
print('Extracting reference patches...')
t0 = time()
patch_size = (7, 7)
data = extract_patches_2d(distorted[:, :height // 2], patch_size)
data = data.reshape(data.shape[0], -1)
data -= np.mean(data, axis=0)
data /= np.std(data, axis=0)
print('done in %.2fs.' % (time() - t0))
###############################################################################
# Learn the dictionary from reference patches
print('Learning the dictionary...')
t0 = time()
dico = MiniBatchDictionaryLearning(n_components=100, alpha=1, n_iter=500)
V = dico.fit(data).components_
dt = time() - t0
print('done in %.2fs.' % dt)
plt.figure(figsize=(4.2, 4))
for i, comp in enumerate(V[:100]):
plt.subplot(10, 10, i + 1)
plt.imshow(comp.reshape(patch_size), cmap=plt.cm.gray_r,
interpolation='nearest')
plt.xticks(())
plt.yticks(())
plt.suptitle('Dictionary learned from Lena patches\n' +
'Train time %.1fs on %d patches' % (dt, len(data)),
fontsize=16)
plt.subplots_adjust(0.08, 0.02, 0.92, 0.85, 0.08, 0.23)
###############################################################################
# Display the distorted image
def show_with_diff(image, reference, title):
"""Helper function to display denoising"""
plt.figure(figsize=(5, 3.3))
plt.subplot(1, 2, 1)
plt.title('Image')
plt.imshow(image, vmin=0, vmax=1, cmap=plt.cm.gray, interpolation='nearest')
plt.xticks(())
plt.yticks(())
plt.subplot(1, 2, 2)
difference = image - reference
plt.title('Difference (norm: %.2f)' % np.sqrt(np.sum(difference ** 2)))
plt.imshow(difference, vmin=-0.5, vmax=0.5, cmap=plt.cm.PuOr,
interpolation='nearest')
plt.xticks(())
plt.yticks(())
plt.suptitle(title, size=16)
plt.subplots_adjust(0.02, 0.02, 0.98, 0.79, 0.02, 0.2)
show_with_diff(distorted, lena, 'Distorted image')
###############################################################################
# Extract noisy patches and reconstruct them using the dictionary
print('Extracting noisy patches... ')
t0 = time()
data = extract_patches_2d(distorted[:, height // 2:], patch_size)
data = data.reshape(data.shape[0], -1)
intercept = np.mean(data, axis=0)
data -= intercept
print('done in %.2fs.' % (time() - t0))
transform_algorithms = [
('Orthogonal Matching Pursuit\n1 atom', 'omp',
{'transform_n_nonzero_coefs': 1}),
('Orthogonal Matching Pursuit\n2 atoms', 'omp',
{'transform_n_nonzero_coefs': 2}),
('Least-angle regression\n5 atoms', 'lars',
{'transform_n_nonzero_coefs': 5}),
('Thresholding\n alpha=0.1', 'threshold', {'transform_alpha': .1})]
reconstructions = {}
for title, transform_algorithm, kwargs in transform_algorithms:
print(title + '...')
reconstructions[title] = lena.copy()
t0 = time()
dico.set_params(transform_algorithm=transform_algorithm, **kwargs)
code = dico.transform(data)
patches = np.dot(code, V)
if transform_algorithm == 'threshold':
patches -= patches.min()
patches /= patches.max()
patches += intercept
patches = patches.reshape(len(data), *patch_size)
if transform_algorithm == 'threshold':
patches -= patches.min()
patches /= patches.max()
reconstructions[title][:, height // 2:] = reconstruct_from_patches_2d(
patches, (width, height // 2))
dt = time() - t0
print('done in %.2fs.' % dt)
show_with_diff(reconstructions[title], lena,
title + ' (time: %.1fs)' % dt)
plt.show()
| bsd-3-clause |
gclenaghan/scikit-learn | examples/preprocessing/plot_function_transformer.py | 158 | 1993 | """
=========================================================
Using FunctionTransformer to select columns
=========================================================
Shows how to use a function transformer in a pipeline. If you know your
dataset's first principle component is irrelevant for a classification task,
you can use the FunctionTransformer to select all but the first column of the
PCA transformed data.
"""
import matplotlib.pyplot as plt
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.decomposition import PCA
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import FunctionTransformer
def _generate_vector(shift=0.5, noise=15):
return np.arange(1000) + (np.random.rand(1000) - shift) * noise
def generate_dataset():
"""
This dataset is two lines with a slope ~ 1, where one has
a y offset of ~100
"""
return np.vstack((
np.vstack((
_generate_vector(),
_generate_vector() + 100,
)).T,
np.vstack((
_generate_vector(),
_generate_vector(),
)).T,
)), np.hstack((np.zeros(1000), np.ones(1000)))
def all_but_first_column(X):
return X[:, 1:]
def drop_first_component(X, y):
"""
Create a pipeline with PCA and the column selector and use it to
transform the dataset.
"""
pipeline = make_pipeline(
PCA(), FunctionTransformer(all_but_first_column),
)
X_train, X_test, y_train, y_test = train_test_split(X, y)
pipeline.fit(X_train, y_train)
return pipeline.transform(X_test), y_test
if __name__ == '__main__':
X, y = generate_dataset()
lw = 0
plt.figure()
plt.scatter(X[:, 0], X[:, 1], c=y, lw=lw)
plt.figure()
X_transformed, y_transformed = drop_first_component(*generate_dataset())
plt.scatter(
X_transformed[:, 0],
np.zeros(len(X_transformed)),
c=y_transformed,
lw=lw,
s=60
)
plt.show()
| bsd-3-clause |
akraft196/pyASC | examples/mplot1.py | 1 | 7267 | #! /usr/bin/env python
#
# quick and dirty processing of the MD All Sky images
from astropy.io import fits
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
from scipy.misc import imsave
import numpy as np
import aplpy
import argparse as ap
import os.path
import logging
import time
def d(ff,box=[]):
#very specific for 16 bit data, since we want to keep the data in uint16
h = fits.open(ff, do_not_scale_image_data=True)
if len(box)==0:
return h[0].header, h[0].data
else:
# figure out 0 vs. 1 based offsets; box is 1 based
return h[0].header, h[0].data[box[1]:box[3], box[0]:box[2]]
def dsum(i0,i1,step = 1, box=[]):
""" for a range of fits files
compute the mean and dispersion from the mean
"""
for i in range(i0,i1+1,step):
ff = 'IMG%05d.FIT' % i
h1, d1 = d(ff,box)
#very specific for 16 bit data, since we want to keep the data in uint16
bzero = h1['BZERO']
bscale = h1['BSCALE']
if i == i0:
sum0 = 1.0
sum1 = d1*bscale+bzero
sum2 = sum1*sum1
#sum1 = d1
#sum2 = d1*d1
h = h1
nx = d1.shape[1]
ny = d1.shape[0]
nz = i1 + 1 - i0
c = np.zeros((nz, ny, nx))
c[0,:,:] = d1.reshape(ny,nx)
else:
sum0 = sum0 + 1.0
sum1 = sum1 + (d1 * bscale + bzero)
sum2 = sum2 + (d1 * bscale + bzero) * (d1 * bscale + bzero)
#sum2 = sum2+d1*d1
c[i - i0,:,:] = d1.reshape(ny,nx)
sum1 = sum1 / sum0
sum2 = sum2 / sum0 - sum1*sum1
print type(sum1), type(sum2)
return h,sum1,np.sqrt(sum2),c
def show(sum):
""" some native matplotlib display,
doesn't show pointsources well at all
"""
ip = plt.imshow(sum)
plt.show()
def show2(sum):
""" aplpy is the better viewer clearly
"""
fig = aplpy.FITSFigure(sum)
#fig.show_grayscale()
fig.show_colorscale()
def show3(sum1,sum2):
""" aplpy is the better viewer clearly
"""
fig = aplpy.FITSFigure(sum1,subplot=(2,2,1))
#fig = aplpy.FITSFigure(sum2,subplot=(2,2,2),figure=1)
fig.show_grayscale()
# For some variations on this theme, e.g. time.time vs. time.clock, see
# http://stackoverflow.com/questions/7370801/measure-time-elapsed-in-python
#
class Dtime(object):
""" Class to help measuring the wall clock time between tagged events
Typical usage:
dt = Dtime()
...
dt.tag('a')
...
dt.tag('b')
"""
def __init__(self, label=".", report=True):
self.start = self.time()
self.init = self.start
self.label = label
self.report = report
self.dtimes = []
dt = self.init - self.init
if self.report:
logging.info("Dtime: %s ADMIT " % self.label + str(self.start))
logging.info("Dtime: %s BEGIN " % self.label + str(dt))
def reset(self, report=True):
self.start = self.time()
self.report = report
self.dtimes = []
def tag(self, mytag):
t0 = self.start
t1 = self.time()
dt = t1 - t0
self.dtimes.append((mytag, dt))
self.start = t1
if self.report:
logging.info("Dtime: %s " % self.label + mytag + " " + str(dt))
return dt
def show(self):
if self.report:
for r in self.dtimes:
logging.info("Dtime: %s " % self.label + str(r[0]) + " " + str(r[1]))
return self.dtimes
def end(self):
t0 = self.init
t1 = self.time()
dt = t1 - t0
if self.report:
logging.info("Dtime: %s END " % self.label + str(dt))
return dt
def time(self):
""" pick the actual OS routine that returns some kind of timer
time.time : wall clock time (include I/O and multitasking overhead)
time.clock : cpu clock time
"""
return np.array([time.clock(), time.time()])
if __name__ == '__main__':
logging.basicConfig(level = logging.INFO)
dt = Dtime("mplot1")
#--start, -s n
#--end, -e n
#--box x1 y1 x2 y2
parser = ap.ArgumentParser(description='Plotting .fits files.')
parser.add_argument('-f', '--frame', nargs = '*', type = int, help = 'Starting and ending parameters for the frames analyzed')
parser.add_argument('-b', '--box', nargs = 4, type = int, help = 'Coordinates for the bottom left corner and top right corner of a rectangle of pixels to be analyzed from the data. In the structure x1, y1, x2, y2 (1 based numbers)')
parser.add_argument('-g', '--graphics', nargs = 1, type = int, default = 0, help = 'Controls whether to display or save graphics. 0: no graphics, 1: display graphics, 2: save graphics as .png')
args = vars(parser.parse_args())
if args['frame'] == None:
count = 0
start = None
end = None
step = 1
#while we have yet to find an end
while end == None:
filename = 'IMG%05d.FIT' % count
#if start has not been found yet, and this file exists
if start == None and os.path.isfile(filename):
start = count
#if start has been found and we finally found a file that doesn't exist, set end to the last file that existed (count - 1.FIT)
elif start != None and not os.path.isfile(filename):
end = count - 1
count += 1
elif len(args['frame']) >= 2 and len(args['frame']) <= 3:
start = args['frame'][0] # starting frame (IMGnnnnn.FIT)
end = args['frame'][1] # ending frame
if len(args['frame']) == 3:
step = args['frame']
else:
step = 1
else:
raise Exception,"-f needs 0, 2, or 3 arguments."
box = args['box'] # BLC and TRC
if box == None:
box = []
dt.tag("start")
# compute the average and dispersion of the series
h1,sum1,sum2,cube = dsum(start,end,step,box=box) # end can be uninitialized here might throw an error?
dt.tag("dsum")
nz = cube.shape[0]
# delta X and Y images
dsumy = sum1 - np.roll(sum1, 1, axis = 0) # change in the y axis
dsumx = sum1 - np.roll(sum1, 1, axis = 1) # change in the x axis
# write them to FITS
fits.writeto('dsumx.fits', dsumx, h1, clobber=True)
fits.writeto('dsumy.fits', dsumy, h1, clobber=True)
fits.writeto('sum1.fits', sum1, h1, clobber=True)
fits.writeto('sum2.fits', sum2, h1, clobber=True)
dt.tag("write2d")
# 3D cube to
h1['NAXIS'] = 3
h1['NAXIS3'] = nz
fits.writeto('cube.fits', cube, h1, clobber=True)
dt.tag("write3d")
if args['graphics'][0] == 1:
# plot the sum1 and sum2 correllation (glueviz should do this)
s1 = sum1.flatten()
s2 = sum2.flatten()
fig = plt.figure()
ax = fig.add_subplot(111)
ax.scatter(s1,s2)
plt.show()
show2(sum1)
show2(sum2)
if args['graphics'][0] == 2:
imsave('sum1.png', sum1)
imsave('sum2.png', sum2)
dt.tag("done")
dt.end()
| mit |
alexsavio/scikit-learn | sklearn/metrics/cluster/bicluster.py | 359 | 2797 | from __future__ import division
import numpy as np
from sklearn.utils.linear_assignment_ import linear_assignment
from sklearn.utils.validation import check_consistent_length, check_array
__all__ = ["consensus_score"]
def _check_rows_and_columns(a, b):
"""Unpacks the row and column arrays and checks their shape."""
check_consistent_length(*a)
check_consistent_length(*b)
checks = lambda x: check_array(x, ensure_2d=False)
a_rows, a_cols = map(checks, a)
b_rows, b_cols = map(checks, b)
return a_rows, a_cols, b_rows, b_cols
def _jaccard(a_rows, a_cols, b_rows, b_cols):
"""Jaccard coefficient on the elements of the two biclusters."""
intersection = ((a_rows * b_rows).sum() *
(a_cols * b_cols).sum())
a_size = a_rows.sum() * a_cols.sum()
b_size = b_rows.sum() * b_cols.sum()
return intersection / (a_size + b_size - intersection)
def _pairwise_similarity(a, b, similarity):
"""Computes pairwise similarity matrix.
result[i, j] is the Jaccard coefficient of a's bicluster i and b's
bicluster j.
"""
a_rows, a_cols, b_rows, b_cols = _check_rows_and_columns(a, b)
n_a = a_rows.shape[0]
n_b = b_rows.shape[0]
result = np.array(list(list(similarity(a_rows[i], a_cols[i],
b_rows[j], b_cols[j])
for j in range(n_b))
for i in range(n_a)))
return result
def consensus_score(a, b, similarity="jaccard"):
"""The similarity of two sets of biclusters.
Similarity between individual biclusters is computed. Then the
best matching between sets is found using the Hungarian algorithm.
The final score is the sum of similarities divided by the size of
the larger set.
Read more in the :ref:`User Guide <biclustering>`.
Parameters
----------
a : (rows, columns)
Tuple of row and column indicators for a set of biclusters.
b : (rows, columns)
Another set of biclusters like ``a``.
similarity : string or function, optional, default: "jaccard"
May be the string "jaccard" to use the Jaccard coefficient, or
any function that takes four arguments, each of which is a 1d
indicator vector: (a_rows, a_columns, b_rows, b_columns).
References
----------
* Hochreiter, Bodenhofer, et. al., 2010. `FABIA: factor analysis
for bicluster acquisition
<https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2881408/>`__.
"""
if similarity == "jaccard":
similarity = _jaccard
matrix = _pairwise_similarity(a, b, similarity)
indices = linear_assignment(1. - matrix)
n_a = len(a[0])
n_b = len(b[0])
return matrix[indices[:, 0], indices[:, 1]].sum() / max(n_a, n_b)
| bsd-3-clause |
plissonf/scikit-learn | sklearn/metrics/tests/test_regression.py | 272 | 6066 | from __future__ import division, print_function
import numpy as np
from itertools import product
from sklearn.utils.testing import assert_raises
from sklearn.utils.testing import assert_equal
from sklearn.utils.testing import assert_almost_equal
from sklearn.utils.testing import assert_array_equal
from sklearn.utils.testing import assert_array_almost_equal
from sklearn.metrics import explained_variance_score
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import mean_squared_error
from sklearn.metrics import median_absolute_error
from sklearn.metrics import r2_score
from sklearn.metrics.regression import _check_reg_targets
def test_regression_metrics(n_samples=50):
y_true = np.arange(n_samples)
y_pred = y_true + 1
assert_almost_equal(mean_squared_error(y_true, y_pred), 1.)
assert_almost_equal(mean_absolute_error(y_true, y_pred), 1.)
assert_almost_equal(median_absolute_error(y_true, y_pred), 1.)
assert_almost_equal(r2_score(y_true, y_pred), 0.995, 2)
assert_almost_equal(explained_variance_score(y_true, y_pred), 1.)
def test_multioutput_regression():
y_true = np.array([[1, 0, 0, 1], [0, 1, 1, 1], [1, 1, 0, 1]])
y_pred = np.array([[0, 0, 0, 1], [1, 0, 1, 1], [0, 0, 0, 1]])
error = mean_squared_error(y_true, y_pred)
assert_almost_equal(error, (1. / 3 + 2. / 3 + 2. / 3) / 4.)
# mean_absolute_error and mean_squared_error are equal because
# it is a binary problem.
error = mean_absolute_error(y_true, y_pred)
assert_almost_equal(error, (1. / 3 + 2. / 3 + 2. / 3) / 4.)
error = r2_score(y_true, y_pred, multioutput='variance_weighted')
assert_almost_equal(error, 1. - 5. / 2)
error = r2_score(y_true, y_pred, multioutput='uniform_average')
assert_almost_equal(error, -.875)
def test_regression_metrics_at_limits():
assert_almost_equal(mean_squared_error([0.], [0.]), 0.00, 2)
assert_almost_equal(mean_absolute_error([0.], [0.]), 0.00, 2)
assert_almost_equal(median_absolute_error([0.], [0.]), 0.00, 2)
assert_almost_equal(explained_variance_score([0.], [0.]), 1.00, 2)
assert_almost_equal(r2_score([0., 1], [0., 1]), 1.00, 2)
def test__check_reg_targets():
# All of length 3
EXAMPLES = [
("continuous", [1, 2, 3], 1),
("continuous", [[1], [2], [3]], 1),
("continuous-multioutput", [[1, 1], [2, 2], [3, 1]], 2),
("continuous-multioutput", [[5, 1], [4, 2], [3, 1]], 2),
("continuous-multioutput", [[1, 3, 4], [2, 2, 2], [3, 1, 1]], 3),
]
for (type1, y1, n_out1), (type2, y2, n_out2) in product(EXAMPLES,
repeat=2):
if type1 == type2 and n_out1 == n_out2:
y_type, y_check1, y_check2, multioutput = _check_reg_targets(
y1, y2, None)
assert_equal(type1, y_type)
if type1 == 'continuous':
assert_array_equal(y_check1, np.reshape(y1, (-1, 1)))
assert_array_equal(y_check2, np.reshape(y2, (-1, 1)))
else:
assert_array_equal(y_check1, y1)
assert_array_equal(y_check2, y2)
else:
assert_raises(ValueError, _check_reg_targets, y1, y2, None)
def test_regression_multioutput_array():
y_true = [[1, 2], [2.5, -1], [4.5, 3], [5, 7]]
y_pred = [[1, 1], [2, -1], [5, 4], [5, 6.5]]
mse = mean_squared_error(y_true, y_pred, multioutput='raw_values')
mae = mean_absolute_error(y_true, y_pred, multioutput='raw_values')
r = r2_score(y_true, y_pred, multioutput='raw_values')
evs = explained_variance_score(y_true, y_pred, multioutput='raw_values')
assert_array_almost_equal(mse, [0.125, 0.5625], decimal=2)
assert_array_almost_equal(mae, [0.25, 0.625], decimal=2)
assert_array_almost_equal(r, [0.95, 0.93], decimal=2)
assert_array_almost_equal(evs, [0.95, 0.93], decimal=2)
# mean_absolute_error and mean_squared_error are equal because
# it is a binary problem.
y_true = [[0, 0]]*4
y_pred = [[1, 1]]*4
mse = mean_squared_error(y_true, y_pred, multioutput='raw_values')
mae = mean_absolute_error(y_true, y_pred, multioutput='raw_values')
r = r2_score(y_true, y_pred, multioutput='raw_values')
assert_array_almost_equal(mse, [1., 1.], decimal=2)
assert_array_almost_equal(mae, [1., 1.], decimal=2)
assert_array_almost_equal(r, [0., 0.], decimal=2)
r = r2_score([[0, -1], [0, 1]], [[2, 2], [1, 1]], multioutput='raw_values')
assert_array_almost_equal(r, [0, -3.5], decimal=2)
assert_equal(np.mean(r), r2_score([[0, -1], [0, 1]], [[2, 2], [1, 1]],
multioutput='uniform_average'))
evs = explained_variance_score([[0, -1], [0, 1]], [[2, 2], [1, 1]],
multioutput='raw_values')
assert_array_almost_equal(evs, [0, -1.25], decimal=2)
# Checking for the condition in which both numerator and denominator is
# zero.
y_true = [[1, 3], [-1, 2]]
y_pred = [[1, 4], [-1, 1]]
r2 = r2_score(y_true, y_pred, multioutput='raw_values')
assert_array_almost_equal(r2, [1., -3.], decimal=2)
assert_equal(np.mean(r2), r2_score(y_true, y_pred,
multioutput='uniform_average'))
evs = explained_variance_score(y_true, y_pred, multioutput='raw_values')
assert_array_almost_equal(evs, [1., -3.], decimal=2)
assert_equal(np.mean(evs), explained_variance_score(y_true, y_pred))
def test_regression_custom_weights():
y_true = [[1, 2], [2.5, -1], [4.5, 3], [5, 7]]
y_pred = [[1, 1], [2, -1], [5, 4], [5, 6.5]]
msew = mean_squared_error(y_true, y_pred, multioutput=[0.4, 0.6])
maew = mean_absolute_error(y_true, y_pred, multioutput=[0.4, 0.6])
rw = r2_score(y_true, y_pred, multioutput=[0.4, 0.6])
evsw = explained_variance_score(y_true, y_pred, multioutput=[0.4, 0.6])
assert_almost_equal(msew, 0.39, decimal=2)
assert_almost_equal(maew, 0.475, decimal=3)
assert_almost_equal(rw, 0.94, decimal=2)
assert_almost_equal(evsw, 0.94, decimal=2)
| bsd-3-clause |
ifuding/Kaggle | PMRCN/Code/siamese_net.py | 1 | 22230 |
from sklearn import *
import sklearn
import pandas as pd
import numpy as np
import xgboost as xgb
import lightgbm as lgb
from time import gmtime, strftime
import numpy.random as rng
from multiprocessing.dummy import Pool
import h5py
import concurrent.futures
import tensorflow as tf
import multiprocessing as mp
from sklearn.cross_validation import KFold
from keras.models import Sequential, Model
from keras.layers.core import Dense, Dropout, Flatten, Reshape
from keras.layers.normalization import BatchNormalization
from keras.layers.embeddings import Embedding
from keras.layers import Input, concatenate, merge
from keras.layers.convolutional import Convolution2D, MaxPooling2D, ZeroPadding2D, AveragePooling2D
from keras.optimizers import SGD, RMSprop, Adam
from keras.callbacks import EarlyStopping
from keras.utils import np_utils
from keras import backend as K
from sklearn.metrics import log_loss
from keras import __version__ as keras_version
graph = tf.get_default_graph()
HIDDEN_UNITS = [64, 16, 8]
DNN_EPOCHS = 40
BATCH_SIZE = 5
DNN_BN = True
DROPOUT_RATE = 0.5
SIAMESE_PAIR_SIZE = 100000
MAX_WORKERS = 8
EMBEDDING_SIZE = 6
full_feature = True
data_folder = '../Data/'
train = pd.read_csv(data_folder + 'training_variants')
#print train.dtypes
test = pd.read_csv(data_folder + 'test_variants')
trainx = pd.read_csv(data_folder + 'training_text', sep="\|\|", engine='python', header=None, skiprows=1, names=["ID","Text"])
#print trainx.dtypes
testx = pd.read_csv(data_folder + 'test_text', sep="\|\|", engine='python', header=None, skiprows=1, names=["ID","Text"])
train = pd.merge(train, trainx, how='left', on='ID').fillna('')
#train = train.iloc[1:1000]
y = train['Class'].values
train = train.drop(['Class'], axis=1)
test = pd.merge(test, testx, how='left', on='ID').fillna('')
pid = test['ID'].values
#df_all = pd.concat((train, test), axis=0, ignore_index=True)
#df_all['Gene_Share'] = df_all.apply(lambda r: sum([1 for w in r['Gene'].split(' ') if w in r['Text'].split(' ')]), axis=1).astype(np.int8)
#df_all['Variation_Share'] = df_all.apply(lambda r: sum([1 for w in r['Variation'].split(' ') if w in r['Text'].split(' ')]), axis=1).astype(np.int8)
#
#print df_all[['Gene_Share', 'Variation_Share']].max()
## exit(0)
#if full_feature:
# #commented for Kaggle Limits
# for i in range(5):
# df_all['Gene_'+str(i)] = df_all['Gene'].map(lambda x: str(x[i]) if len(x)>i else '')
# df_all['Variation'+str(i)] = df_all['Variation'].map(lambda x: str(x[i]) if len(x)>i else '')
# print df_all.dtypes
#
# gen_var_lst = sorted(list(train.Gene.unique()) + list(train.Variation.unique()))
# print(len(gen_var_lst))
# gen_var_lst = [x for x in gen_var_lst if len(x.split(' '))==1]
# print(len(gen_var_lst))
# i_ = 0
# #commented for Kaggle Limits
# for gen_var_lst_itm in gen_var_lst:
# if i_ % 100 == 0: print(i_)
# df_all['GV_'+str(gen_var_lst_itm)] = df_all['Text'].map(lambda x: str(x).count(str(gen_var_lst_itm))).astype(np.int8)
# i_ += 1
# if i_ == 5:
# break
#
#for c in df_all.columns:
# if df_all[c].dtype == 'object':
# if c in ['Gene','Variation']:
# lbl = preprocessing.LabelEncoder()
# df_all[c+'_lbl_enc'] = lbl.fit_transform(df_all[c].values)
# df_all[c+'_len'] = df_all[c].map(lambda x: len(str(x)))
# df_all[c+'_words'] = df_all[c].map(lambda x: len(str(x).split(' ')))
# elif c != 'Text':
# lbl = preprocessing.LabelEncoder()
# df_all[c] = lbl.fit_transform(df_all[c].values)
# if c=='Text':
# df_all[c+'_len'] = df_all[c].map(lambda x: len(str(x)))
# df_all[c+'_words'] = df_all[c].map(lambda x: len(str(x).split(' ')))
#
#train = df_all.iloc[:len(train)]
#print "... train dtypes before svd ..."
#print train.dtypes
#print train.head()
#exit(0)
#test = df_all.iloc[len(train):]
#
#class cust_regression_vals(sklearn.base.BaseEstimator, sklearn.base.TransformerMixin):
# def fit(self, x, y=None):
# return self
# def transform(self, x):
# x = x.drop(['Gene', 'Variation','ID','Text'],axis=1).values
# return x
#
#class cust_txt_col(sklearn.base.BaseEstimator, sklearn.base.TransformerMixin):
# def __init__(self, key):
# self.key = key
# def fit(self, x, y=None):
# return self
# def transform(self, x):
# return x[self.key].apply(str)
#
#print('Pipeline...')
#fp = pipeline.Pipeline([
# ('union', pipeline.FeatureUnion(
# n_jobs = -1,
# transformer_list = [
# ('standard', cust_regression_vals()),
# ('pi1', pipeline.Pipeline([('Gene', cust_txt_col('Gene')), ('count_Gene', feature_extraction.text.CountVectorizer(analyzer=u'char', ngram_range=(1, 8))), ('tsvd1', decomposition.TruncatedSVD(n_components=20, n_iter=25, random_state=12))])),
# ('pi2', pipeline.Pipeline([('Variation', cust_txt_col('Variation')), ('count_Variation', feature_extraction.text.CountVectorizer(analyzer=u'char', ngram_range=(1, 8))), ('tsvd2', decomposition.TruncatedSVD(n_components=20, n_iter=25, random_state=12))])),
# #commented for Kaggle Limits
# ('pi3', pipeline.Pipeline([('Text', cust_txt_col('Text')), ('tfidf_Text', feature_extraction.text.TfidfVectorizer(ngram_range=(1, 2))), ('tsvd3', decomposition.TruncatedSVD(n_components=50, n_iter=25, random_state=12))]))
# ])
# )])
#
#train = fp.fit_transform(train);
#print type(train)
#print(train.shape)
#print (train.nbytes)
#np.save("train_array", train)
## print(df.dtypes)
## print(df.memory_usage())
#test = fp.transform(test); print(test.shape)
#np.save("test_array", test)
#exit(0)
train = np.load("./train_array.npy")
test = np.load("./test_array.npy")
# siamese_features_array = np.load("./siamese_features_array_2017_09_15_07_57_44.npy")
y = y - 1 #fix for zero bound array
CONTINUOUS_INDICES = []
SPARSE_INDICES = []
for i in range((train.shape)[1]):
if (i >= 3205 and i <= 3212):
pass
elif (i >= 2 and i <= 113): # or (i >= 114 and i <= 3204):
SPARSE_INDICES.append(i)
else:
CONTINUOUS_INDICES.append(i)
#train = train[:, CONTINUOUS_INDICES]
#test = test[:, CONTINUOUS_INDICES]
print('train shape after loading and selecting trainging columns: %s' % str(train.shape))
siamese_train_len = len(train) // 3
print('siamese_train_len is %d' % (siamese_train_len))
siamese_train_data = train[:siamese_train_len]
siamese_train_label = y[:siamese_train_len]
lgbm_train_data = train[siamese_train_len:]
lgbm_train_label = y[siamese_train_len:]
#train = train[:200]
#y = y[:200]
#test = test[:200]
#pid = pid[:200]
def xgbTrain(train_data, train_label, flod = 5):
"""
"""
denom = 0
fold = 5 #Change to 5, 1 for Kaggle Limits
models = []
for i in range(fold):
params = {
'eta': 0.03333,
'max_depth': 4,
'objective': 'multi:softprob',
'eval_metric': 'mlogloss',
'num_class': 9,
'seed': i,
'silent': True
}
x1, x2, y1, y2 = model_selection.train_test_split(train_data, train_label, test_size=0.18, random_state=i)
watchlist = [(xgb.DMatrix(x1, y1), 'train'), (xgb.DMatrix(x2, y2), 'valid')]
model = xgb.train(params, xgb.DMatrix(x1, y1), 1000, watchlist, verbose_eval=50, early_stopping_rounds=100)
score1 = metrics.log_loss(y2, model.predict(xgb.DMatrix(x2), ntree_limit=model.best_ntree_limit), labels = list(range(9)))
#print(score1)
models.append((model, 'x'))
return models
def lgbm_train(train_data, train_label, fold = 5):
"""
LGB Training
"""
# print train.shape
# print siamese_features_array.shape
# train_merge = siamese_features_array #np.concatenate((train, siamese_features_array), axis = 1)
# print train_merge.shape
# # exit(0)
print("Over all training size:")
print(train_data.shape)
# train_data = train_merge#[:train_len * 3 / 10]
# train_label = lgbm_train_label#[:train_len * 3 / 10]
#valide_data = train_merge[train_len * 9 / 10:]
#valide_label = y[train_len * 9 / 10:]
models = []
for i in range(fold):
d_train = lgb.Dataset(train_data, train_label) #, categorical_feature = SPARCE_INDICES)
#d_valide = lgb.Dataset(valide_data, valide_label)
params = {
'task': 'train',
'boosting_type': 'gbdt',
'objective': 'multiclass',
'metric': {'multi_logloss'},
'num_class': 9,
# 'num_leaves': 256,
# 'max_depth': 12,
# 'feature_fraction': 0.9,
# 'bagging_fraction': 0.95,
# 'bagging_freq': 5,
'num_leaves': 60, # 60,
# 'min_sum_hessian_in_leaf': 20,
'max_depth': 10, # 10,
'learning_rate': 0.02, # 0.02,
'feature_fraction': 0.5,
'verbose': 0,
# 'valid_sets': [d_valide],
'num_boost_round': 327,
'feature_fraction_seed': i,
# 'bagging_fraction': 0.9,
# 'bagging_freq': 15,
# 'bagging_seed': i,
# 'early_stopping_round': 10
# 'random_state': 10
# 'verbose_eval': 20
#'min_data_in_leaf': 665
}
# ROUNDS = 1
print('fold: %d th light GBM train :-)' % (i))
# params['feature_fraction_seed'] = i
#bst = lgb.train(
# params ,
# d_train,
# verbose_eval = False
# # valid_sets = [d_valide]
# #num_boost_round = 1
# )
cv_result = lgb.cv(params, d_train, nfold=10)
pd.DataFrame(cv_result).to_csv('cv_result', index = False)
exit(0)
# pred = model_eval(bst, 'l', test)
#print pred.shape
#print pred[0, :]
models.append((bst, 'l'))
return models
def create_model(input_len):
model = Sequential()
model.add(Dense(HIDDEN_UNITS[0], activation='sigmoid', input_dim = input_len))
if DNN_BN:
model.add(BatchNormalization())
if DROPOUT_RATE > 0:
model.add(Dropout(DROPOUT_RATE))
model.add(Dense(HIDDEN_UNITS[1], activation='sigmoid'))
if DNN_BN:
model.add(BatchNormalization())
if DROPOUT_RATE > 0:
model.add(Dropout(DROPOUT_RATE))
# model.add(Dropout(0.1))
#model.add(Dense(32, activation='relu'))
#model.add(Dropout(0.2))
model.add(Dense(9, activation='softmax'))
# optimizer = SGD(lr=1e-3, decay=1e-6, momentum=0.9, nesterov=True)
optimizer = RMSprop(lr=1e-3, rho = 0.9, epsilon = 1e-8)
model.compile(optimizer=optimizer, loss='categorical_crossentropy', metrics = ['accuracy'])
return model
def create_embedding_model(CONTINUE_SIZE, SPARSE_SIZE):
"""
"""
print('CONTINUOUS_SIZE = %d' % CONTINUE_SIZE)
print('SPARSE_SIZE = %d' % SPARSE_SIZE)
sparse_feature = Input(shape=(SPARSE_SIZE,))
sparse_embedding = Embedding(55, EMBEDDING_SIZE, input_length = SPARSE_SIZE)(sparse_feature)
sparse_embedding = Reshape((EMBEDDING_SIZE * SPARSE_SIZE,))(sparse_embedding)
# print "model input size: %d" % CONTINUOUS_COLUMNS
dense_input = Input(shape=(CONTINUE_SIZE,))
merge_input = concatenate([dense_input, sparse_embedding], axis = 1)
merge_len = CONTINUE_SIZE + EMBEDDING_SIZE * SPARSE_SIZE
output = create_model(merge_len)(merge_input)
model = Model([dense_input, sparse_feature], output)
optimizer = RMSprop(lr=1e-3, rho = 0.9, epsilon = 1e-8)
# optimizer = SGD(lr=1e-2, decay=1e-6, momentum=0.9, nesterov=True)
model.compile(optimizer = Adam(),
loss='categorical_crossentropy', metrics = ['accuracy'])
return model
def keras_train(train_data, train_target, nfolds = 10):
"""
Detect Fish or noFish
"""
print("Start gen training data, shuffle and normalize!")
#train_data = train
train_target = np_utils.to_categorical(train_target)
# train_data, train_target, siamese_data_loader = siamese_train(siamese_train_data, siamese_train_label)
kf = KFold(len(train_target), n_folds=nfolds, shuffle=True)
num_fold = 0
models = []
for train_index, test_index in kf:
# model = create_model(classes = 2)
model = create_embedding_model(len(CONTINUOUS_INDICES), len(SPARSE_INDICES))
# model = create_siamese_net((train.shape)[1])
X_train = train_data[train_index]
Y_train = train_target[train_index]
print('Positive samples in train: %d' % np.sum(Y_train))
print('Negative samples in train: %d' % (len(Y_train) - np.sum(Y_train)))
X_valid = train_data[test_index]
Y_valid = train_target[test_index]
print('Positive samples in valide: %d' % np.sum(Y_valid))
print('Negative samples in valide: %d' % (len(Y_valid) - np.sum(Y_valid)))
num_fold += 1
print('Start KFold number {} from {}'.format(num_fold, nfolds))
print('Split train: ', len(X_train), len(Y_train))
print('Split valid: ', len(X_valid), len(Y_valid))
callbacks = [
EarlyStopping(monitor='val_loss', patience=5, verbose=0),
]
model.fit([X_train[:, CONTINUOUS_INDICES], X_train[:, SPARSE_INDICES]],
Y_train, batch_size=BATCH_SIZE, epochs=DNN_EPOCHS,
shuffle=True, verbose=2,
validation_data=([X_valid[:, CONTINUOUS_INDICES], X_valid[:, SPARSE_INDICES]], Y_valid)
, callbacks=callbacks)
model_name = 'keras' + strftime('_%Y_%m_%d_%H_%M_%S', gmtime())
#model.save_weights(model_name)
#siamese_features_array = gen_siamese_features(model, lgbm_train_data, siamese_train_data, siamese_train_label)
models.append((model, 'k'))
break
return models #, siamese_features_array
def model_eval(model, model_type, data_frame):
"""
"""
if model_type == 'l':
preds = model.predict(data_frame)
elif model_type == 'k':
preds = model.predict(data_frame, batch_size=BATCH_SIZE, verbose=2)
elif model_type == 't':
print("ToDO")
elif model_type == 'x':
preds = model.predict(xgb.DMatrix(data_frame), ntree_limit=model.best_ntree_limit+80)
return preds
def gen_sub(models, merge_features):
"""
Evaluate single Type model
"""
print('Start generate submission!')
preds = None
for (model, model_type) in models:
pred = model_eval(model, model_type, merge_features)
#print pred.shape
#print pred[0, :]
if preds is None:
preds = pred.copy()
else:
preds += pred
preds /= len(models)
submission = pd.DataFrame(preds, columns=['class'+str(c+1) for c in range(9)])
submission['ID'] = pid
sub_name = "submission" + strftime('_%Y_%m_%d_%H_%M_%S', gmtime()) + ".csv"
print('Output to ' + sub_name)
submission.to_csv(sub_name, index=False)
def create_siamese_net(input_size):
"""
"""
input_shape = (input_size, )
left_input = Input(input_shape)
right_input = Input(input_shape)
#build model to use in each siamese 'leg'
model = Sequential()
model.add(Dense(HIDDEN_UNITS[0], activation='sigmoid', input_dim = input_size))
if DNN_BN:
model.add(BatchNormalization())
if DROPOUT_RATE > 0:
model.add(Dropout(DROPOUT_RATE))
model.add(Dense(HIDDEN_UNITS[1], activation='sigmoid'))
if DNN_BN:
model.add(BatchNormalization())
if DROPOUT_RATE > 0:
model.add(Dropout(DROPOUT_RATE))
#encode each of the two inputs into a vector with the convnet
encoded_l = model(left_input)
encoded_r = model(right_input)
#merge two encoded inputs with the l1 distance between them
L1_distance = lambda x: K.abs(x[0]-x[1])
both = merge([encoded_l,encoded_r], mode = L1_distance, output_shape=lambda x: x[0])
merge_layer = Dense(HIDDEN_UNITS[2],activation='sigmoid')(both)
prediction = Dense(1,activation='sigmoid')(merge_layer)
siamese_net = Model(input=[left_input,right_input],output=prediction)
#optimizer = SGD(0.0004,momentum=0.6,nesterov=True,decay=0.0003)
optimizer = Adam()
#//TODO: get layerwise learning rates and momentum annealing scheme described in paperworking
siamese_net.compile(loss="binary_crossentropy",optimizer=optimizer)
# print siamese_net.count_params()
return siamese_net
class Siamese_Loader:
#For loading batches and testing tasks to a siamese net
def __init__(self,Xtrain,Xval = None):
self.Xval = Xval
self.Xtrain = Xtrain
self.n_classes = Xtrain.shape[0]
self.feature_size = (Xtrain[0].shape)[1]
self.n_examples = np.array([x.shape[0] for x in Xtrain])
self.n_tot_examples = np.sum(self.n_examples)
print('examples of different classes: %s' % str(self.n_examples))
# self.n_val,self.n_ex_val,_,_ = Xval.shape
def get_batch(self,n):
#Create batch of pairs, half same class, half different class
categories = rng.choice(self.n_classes,size=(n,),replace=True)
pairs=np.zeros((2, n, self.feature_size))
targets=np.zeros((n,))
positive_begin_pos = n * 1 // 2
targets[positive_begin_pos:] = 1
categories_list = []
for i in range(n):
category = categories[i]
idx_1 = rng.randint(0, self.n_examples[category])
pairs[0][i] = self.Xtrain[category][idx_1] #.reshape(self.feature_size)
#pick images of same class for 1st half, different for 2nd
category_2 = category if i >= positive_begin_pos else (category + rng.randint(1,self.n_classes)) % self.n_classes
idx_2 = rng.randint(0,self.n_examples[category_2])
while i >= positive_begin_pos and idx_2 == idx_1:
idx_2 = rng.randint(0,self.n_examples[category_2])
pairs[1][i] = self.Xtrain[category_2][idx_2] #.reshape(self.w,self.h,1)
categories_list.append((category, category_2))
#pd.DataFrame(categories_list).to_csv('categories', index=False)
#exit(0)
# shuflle pairs to mix positive and negative
rng.shuffle(pairs)
return pairs, targets
def gen_test_on_support_data(Xsupport, Xtest):
"""
"""
n_support, feature_size = Xsupport.shape
pairs = np.zeros((2, n_support, feature_size))
pairs[0] = Xtest
pairs[1] = Xsupport
return list(pairs)
def siamese_train(siamese_train_data, siamese_train_label):
"""
"""
train_data = [[] for i in range(9)]
label_ind = 0
for feature in siamese_train_data:
train_data[siamese_train_label[label_ind]].append(feature)
label_ind += 1
train_data = np.array([np.array(xi) for xi in train_data])
print("train data shape before gen pair")
print(train_data.shape)
siamese_data_loader = Siamese_Loader(train_data, test)
pairs, targets = siamese_data_loader.get_batch(SIAMESE_PAIR_SIZE)
return pairs, targets, siamese_data_loader
def gen_siamese_features_meta(model, Xsupport_label, Xsupport, Xtest):
"""
"""
siamese_pair = gen_test_on_support_data(Xsupport, Xtest)
global graph
with graph.as_default():
preds = model.predict(siamese_pair, batch_size=BATCH_SIZE, verbose=2)
preds = np.insert(preds, 1, Xsupport_label, axis = 1)
preds = pd.DataFrame(preds, columns = ['sim', 'class'])
siamese_features = preds.groupby('class', sort = False) \
.agg({'sim': ['max', 'min', 'median', 'mean', 'std']})
max_class = siamese_features['sim']['max'].idxmax()
siamese_features = np.insert(siamese_features.values.flatten(), 0, max_class, axis = 0)
return siamese_features
def gen_siamese_features(siamese_model, Xtest, Xsupport, Xsupport_label):
"""
"""
if MAX_WORKERS <= 0:
print("MAX_WORKERS should >= 1", file=sys.stderr)
exit(1)
siamese_features_array = list(range(len(Xtest)))
test_begin = 0
while test_begin < len(Xtest):
test_end = min(test_begin + MAX_WORKERS, len(Xtest))
with concurrent.futures.ThreadPoolExecutor(max_workers = MAX_WORKERS) as executor:
future_predict = {executor.submit(gen_siamese_features_meta, siamese_model,
Xsupport_label,
Xsupport,
Xtest[ind]): ind for ind in range(test_begin, test_end)}
for future in concurrent.futures.as_completed(future_predict):
ind = future_predict[future]
try:
siamese_features = future.result()
siamese_features_array[ind] = siamese_features
except Exception as exc:
print('%dth feature generated an exception: %s' % (ind, exc))
test_begin = test_end
if test_begin % 100 == 0:
print('Gen %d siamsese features' % test_begin)
if test_begin != len(Xtest):
print("Only gen %d siamese features" % test_begin, file=sys.stderr)
exit(1)
siamese_features_array = np.array(siamese_features_array)
pd.DataFrame(siamese_features_array[:, 0]).astype(np.int8).to_csv('pred_label', index = False)
return siamese_features_array
if __name__ == "__main__":
model_k = keras_train(train, y, 10)
#np.save("siamese_features_array" + \
# strftime('_%Y_%m_%d_%H_%M_%S', gmtime()) , siamese_features_array)
# gen_sub(model_k, 'k', th, F1)
# ind = np.array([i * 5 for i in range(9)])
# xgbTrain(siamese_features_array[:, ind], lgbm_train_label);
#lgbm_features = siamese_features_array #np.concatenate((lgbm_train_data, siamese_features_array),
# model_l = lgbm_train(train, y, 10) #lgbm_features, lgbm_train_label, 10)#model_k)
# siamese_features_test_array = siamese_test(model_k[0][0], test)
#np.save("siamese_features_test_array" + \
# strftime('_%Y_%m_%d_%H_%M_%S', gmtime()) , siamese_features_test_array)
##model_x = xgbTrain(5)#model_k)
#gen_sub(model_l, siamese_features_test_array) #model_k)
| apache-2.0 |
lhilt/scipy | scipy/stats/tests/test_morestats.py | 4 | 70469 | # Author: Travis Oliphant, 2002
#
# Further enhancements and tests added by numerous SciPy developers.
#
from __future__ import division, print_function, absolute_import
import warnings
import numpy as np
from numpy.random import RandomState
from numpy.testing import (assert_array_equal,
assert_almost_equal, assert_array_less, assert_array_almost_equal,
assert_, assert_allclose, assert_equal, assert_warns)
import pytest
from pytest import raises as assert_raises
from scipy._lib._numpy_compat import suppress_warnings
from scipy import stats
from .common_tests import check_named_results
# Matplotlib is not a scipy dependency but is optionally used in probplot, so
# check if it's available
try:
import matplotlib
matplotlib.rcParams['backend'] = 'Agg'
import matplotlib.pyplot as plt
have_matplotlib = True
except Exception:
have_matplotlib = False
# test data gear.dat from NIST for Levene and Bartlett test
# https://www.itl.nist.gov/div898/handbook/eda/section3/eda3581.htm
g1 = [1.006, 0.996, 0.998, 1.000, 0.992, 0.993, 1.002, 0.999, 0.994, 1.000]
g2 = [0.998, 1.006, 1.000, 1.002, 0.997, 0.998, 0.996, 1.000, 1.006, 0.988]
g3 = [0.991, 0.987, 0.997, 0.999, 0.995, 0.994, 1.000, 0.999, 0.996, 0.996]
g4 = [1.005, 1.002, 0.994, 1.000, 0.995, 0.994, 0.998, 0.996, 1.002, 0.996]
g5 = [0.998, 0.998, 0.982, 0.990, 1.002, 0.984, 0.996, 0.993, 0.980, 0.996]
g6 = [1.009, 1.013, 1.009, 0.997, 0.988, 1.002, 0.995, 0.998, 0.981, 0.996]
g7 = [0.990, 1.004, 0.996, 1.001, 0.998, 1.000, 1.018, 1.010, 0.996, 1.002]
g8 = [0.998, 1.000, 1.006, 1.000, 1.002, 0.996, 0.998, 0.996, 1.002, 1.006]
g9 = [1.002, 0.998, 0.996, 0.995, 0.996, 1.004, 1.004, 0.998, 0.999, 0.991]
g10 = [0.991, 0.995, 0.984, 0.994, 0.997, 0.997, 0.991, 0.998, 1.004, 0.997]
class TestBayes_mvs(object):
def test_basic(self):
# Expected values in this test simply taken from the function. For
# some checks regarding correctness of implementation, see review in
# gh-674
data = [6, 9, 12, 7, 8, 8, 13]
mean, var, std = stats.bayes_mvs(data)
assert_almost_equal(mean.statistic, 9.0)
assert_allclose(mean.minmax, (7.1036502226125329, 10.896349777387467),
rtol=1e-14)
assert_almost_equal(var.statistic, 10.0)
assert_allclose(var.minmax, (3.1767242068607087, 24.45910381334018),
rtol=1e-09)
assert_almost_equal(std.statistic, 2.9724954732045084, decimal=14)
assert_allclose(std.minmax, (1.7823367265645145, 4.9456146050146312),
rtol=1e-14)
def test_empty_input(self):
assert_raises(ValueError, stats.bayes_mvs, [])
def test_result_attributes(self):
x = np.arange(15)
attributes = ('statistic', 'minmax')
res = stats.bayes_mvs(x)
for i in res:
check_named_results(i, attributes)
class TestMvsdist(object):
def test_basic(self):
data = [6, 9, 12, 7, 8, 8, 13]
mean, var, std = stats.mvsdist(data)
assert_almost_equal(mean.mean(), 9.0)
assert_allclose(mean.interval(0.9), (7.1036502226125329,
10.896349777387467), rtol=1e-14)
assert_almost_equal(var.mean(), 10.0)
assert_allclose(var.interval(0.9), (3.1767242068607087,
24.45910381334018), rtol=1e-09)
assert_almost_equal(std.mean(), 2.9724954732045084, decimal=14)
assert_allclose(std.interval(0.9), (1.7823367265645145,
4.9456146050146312), rtol=1e-14)
def test_empty_input(self):
assert_raises(ValueError, stats.mvsdist, [])
def test_bad_arg(self):
# Raise ValueError if fewer than two data points are given.
data = [1]
assert_raises(ValueError, stats.mvsdist, data)
def test_warns(self):
# regression test for gh-5270
# make sure there are no spurious divide-by-zero warnings
with warnings.catch_warnings():
warnings.simplefilter('error', RuntimeWarning)
[x.mean() for x in stats.mvsdist([1, 2, 3])]
[x.mean() for x in stats.mvsdist([1, 2, 3, 4, 5])]
class TestShapiro(object):
def test_basic(self):
x1 = [0.11, 7.87, 4.61, 10.14, 7.95, 3.14, 0.46,
4.43, 0.21, 4.75, 0.71, 1.52, 3.24,
0.93, 0.42, 4.97, 9.53, 4.55, 0.47, 6.66]
w, pw = stats.shapiro(x1)
assert_almost_equal(w, 0.90047299861907959, 6)
assert_almost_equal(pw, 0.042089745402336121, 6)
x2 = [1.36, 1.14, 2.92, 2.55, 1.46, 1.06, 5.27, -1.11,
3.48, 1.10, 0.88, -0.51, 1.46, 0.52, 6.20, 1.69,
0.08, 3.67, 2.81, 3.49]
w, pw = stats.shapiro(x2)
assert_almost_equal(w, 0.9590270, 6)
assert_almost_equal(pw, 0.52460, 3)
# Verified against R
np.random.seed(12345678)
x3 = stats.norm.rvs(loc=5, scale=3, size=100)
w, pw = stats.shapiro(x3)
assert_almost_equal(w, 0.9772805571556091, decimal=6)
assert_almost_equal(pw, 0.08144091814756393, decimal=3)
# Extracted from original paper
x4 = [0.139, 0.157, 0.175, 0.256, 0.344, 0.413, 0.503, 0.577, 0.614,
0.655, 0.954, 1.392, 1.557, 1.648, 1.690, 1.994, 2.174, 2.206,
3.245, 3.510, 3.571, 4.354, 4.980, 6.084, 8.351]
W_expected = 0.83467
p_expected = 0.000914
w, pw = stats.shapiro(x4)
assert_almost_equal(w, W_expected, decimal=4)
assert_almost_equal(pw, p_expected, decimal=5)
def test_2d(self):
x1 = [[0.11, 7.87, 4.61, 10.14, 7.95, 3.14, 0.46,
4.43, 0.21, 4.75], [0.71, 1.52, 3.24,
0.93, 0.42, 4.97, 9.53, 4.55, 0.47, 6.66]]
w, pw = stats.shapiro(x1)
assert_almost_equal(w, 0.90047299861907959, 6)
assert_almost_equal(pw, 0.042089745402336121, 6)
x2 = [[1.36, 1.14, 2.92, 2.55, 1.46, 1.06, 5.27, -1.11,
3.48, 1.10], [0.88, -0.51, 1.46, 0.52, 6.20, 1.69,
0.08, 3.67, 2.81, 3.49]]
w, pw = stats.shapiro(x2)
assert_almost_equal(w, 0.9590270, 6)
assert_almost_equal(pw, 0.52460, 3)
def test_empty_input(self):
assert_raises(ValueError, stats.shapiro, [])
assert_raises(ValueError, stats.shapiro, [[], [], []])
def test_not_enough_values(self):
assert_raises(ValueError, stats.shapiro, [1, 2])
assert_raises(ValueError, stats.shapiro, [[], [2]])
def test_bad_arg(self):
# Length of x is less than 3.
x = [1]
assert_raises(ValueError, stats.shapiro, x)
def test_nan_input(self):
x = np.arange(10.)
x[9] = np.nan
w, pw = stats.shapiro(x)
assert_equal(w, np.nan)
assert_almost_equal(pw, 1.0)
class TestAnderson(object):
def test_normal(self):
rs = RandomState(1234567890)
x1 = rs.standard_exponential(size=50)
x2 = rs.standard_normal(size=50)
A, crit, sig = stats.anderson(x1)
assert_array_less(crit[:-1], A)
A, crit, sig = stats.anderson(x2)
assert_array_less(A, crit[-2:])
v = np.ones(10)
v[0] = 0
A, crit, sig = stats.anderson(v)
# The expected statistic 3.208057 was computed independently of scipy.
# For example, in R:
# > library(nortest)
# > v <- rep(1, 10)
# > v[1] <- 0
# > result <- ad.test(v)
# > result$statistic
# A
# 3.208057
assert_allclose(A, 3.208057)
def test_expon(self):
rs = RandomState(1234567890)
x1 = rs.standard_exponential(size=50)
x2 = rs.standard_normal(size=50)
A, crit, sig = stats.anderson(x1, 'expon')
assert_array_less(A, crit[-2:])
olderr = np.seterr(all='ignore')
try:
A, crit, sig = stats.anderson(x2, 'expon')
finally:
np.seterr(**olderr)
assert_(A > crit[-1])
def test_gumbel(self):
# Regression test for gh-6306. Before that issue was fixed,
# this case would return a2=inf.
v = np.ones(100)
v[0] = 0.0
a2, crit, sig = stats.anderson(v, 'gumbel')
# A brief reimplementation of the calculation of the statistic.
n = len(v)
xbar, s = stats.gumbel_l.fit(v)
logcdf = stats.gumbel_l.logcdf(v, xbar, s)
logsf = stats.gumbel_l.logsf(v, xbar, s)
i = np.arange(1, n+1)
expected_a2 = -n - np.mean((2*i - 1) * (logcdf + logsf[::-1]))
assert_allclose(a2, expected_a2)
def test_bad_arg(self):
assert_raises(ValueError, stats.anderson, [1], dist='plate_of_shrimp')
def test_result_attributes(self):
rs = RandomState(1234567890)
x = rs.standard_exponential(size=50)
res = stats.anderson(x)
attributes = ('statistic', 'critical_values', 'significance_level')
check_named_results(res, attributes)
def test_gumbel_l(self):
# gh-2592, gh-6337
# Adds support to 'gumbel_r' and 'gumbel_l' as valid inputs for dist.
rs = RandomState(1234567890)
x = rs.gumbel(size=100)
A1, crit1, sig1 = stats.anderson(x, 'gumbel')
A2, crit2, sig2 = stats.anderson(x, 'gumbel_l')
assert_allclose(A2, A1)
def test_gumbel_r(self):
# gh-2592, gh-6337
# Adds support to 'gumbel_r' and 'gumbel_l' as valid inputs for dist.
rs = RandomState(1234567890)
x1 = rs.gumbel(size=100)
x2 = np.ones(100)
A1, crit1, sig1 = stats.anderson(x1, 'gumbel_r')
A2, crit2, sig2 = stats.anderson(x2, 'gumbel_r')
assert_array_less(A1, crit1[-2:])
assert_(A2 > crit2[-1])
class TestAndersonKSamp(object):
def test_example1a(self):
# Example data from Scholz & Stephens (1987), originally
# published in Lehmann (1995, Nonparametrics, Statistical
# Methods Based on Ranks, p. 309)
# Pass a mixture of lists and arrays
t1 = [38.7, 41.5, 43.8, 44.5, 45.5, 46.0, 47.7, 58.0]
t2 = np.array([39.2, 39.3, 39.7, 41.4, 41.8, 42.9, 43.3, 45.8])
t3 = np.array([34.0, 35.0, 39.0, 40.0, 43.0, 43.0, 44.0, 45.0])
t4 = np.array([34.0, 34.8, 34.8, 35.4, 37.2, 37.8, 41.2, 42.8])
Tk, tm, p = stats.anderson_ksamp((t1, t2, t3, t4), midrank=False)
assert_almost_equal(Tk, 4.449, 3)
assert_array_almost_equal([0.4985, 1.3237, 1.9158, 2.4930, 3.2459],
tm[0:5], 4)
assert_allclose(p, 0.0021, atol=0.00025)
def test_example1b(self):
# Example data from Scholz & Stephens (1987), originally
# published in Lehmann (1995, Nonparametrics, Statistical
# Methods Based on Ranks, p. 309)
# Pass arrays
t1 = np.array([38.7, 41.5, 43.8, 44.5, 45.5, 46.0, 47.7, 58.0])
t2 = np.array([39.2, 39.3, 39.7, 41.4, 41.8, 42.9, 43.3, 45.8])
t3 = np.array([34.0, 35.0, 39.0, 40.0, 43.0, 43.0, 44.0, 45.0])
t4 = np.array([34.0, 34.8, 34.8, 35.4, 37.2, 37.8, 41.2, 42.8])
Tk, tm, p = stats.anderson_ksamp((t1, t2, t3, t4), midrank=True)
assert_almost_equal(Tk, 4.480, 3)
assert_array_almost_equal([0.4985, 1.3237, 1.9158, 2.4930, 3.2459],
tm[0:5], 4)
assert_allclose(p, 0.0020, atol=0.00025)
def test_example2a(self):
# Example data taken from an earlier technical report of
# Scholz and Stephens
# Pass lists instead of arrays
t1 = [194, 15, 41, 29, 33, 181]
t2 = [413, 14, 58, 37, 100, 65, 9, 169, 447, 184, 36, 201, 118]
t3 = [34, 31, 18, 18, 67, 57, 62, 7, 22, 34]
t4 = [90, 10, 60, 186, 61, 49, 14, 24, 56, 20, 79, 84, 44, 59, 29,
118, 25, 156, 310, 76, 26, 44, 23, 62]
t5 = [130, 208, 70, 101, 208]
t6 = [74, 57, 48, 29, 502, 12, 70, 21, 29, 386, 59, 27]
t7 = [55, 320, 56, 104, 220, 239, 47, 246, 176, 182, 33]
t8 = [23, 261, 87, 7, 120, 14, 62, 47, 225, 71, 246, 21, 42, 20, 5,
12, 120, 11, 3, 14, 71, 11, 14, 11, 16, 90, 1, 16, 52, 95]
t9 = [97, 51, 11, 4, 141, 18, 142, 68, 77, 80, 1, 16, 106, 206, 82,
54, 31, 216, 46, 111, 39, 63, 18, 191, 18, 163, 24]
t10 = [50, 44, 102, 72, 22, 39, 3, 15, 197, 188, 79, 88, 46, 5, 5, 36,
22, 139, 210, 97, 30, 23, 13, 14]
t11 = [359, 9, 12, 270, 603, 3, 104, 2, 438]
t12 = [50, 254, 5, 283, 35, 12]
t13 = [487, 18, 100, 7, 98, 5, 85, 91, 43, 230, 3, 130]
t14 = [102, 209, 14, 57, 54, 32, 67, 59, 134, 152, 27, 14, 230, 66,
61, 34]
Tk, tm, p = stats.anderson_ksamp((t1, t2, t3, t4, t5, t6, t7, t8,
t9, t10, t11, t12, t13, t14),
midrank=False)
assert_almost_equal(Tk, 3.288, 3)
assert_array_almost_equal([0.5990, 1.3269, 1.8052, 2.2486, 2.8009],
tm[0:5], 4)
assert_allclose(p, 0.0041, atol=0.00025)
def test_example2b(self):
# Example data taken from an earlier technical report of
# Scholz and Stephens
t1 = [194, 15, 41, 29, 33, 181]
t2 = [413, 14, 58, 37, 100, 65, 9, 169, 447, 184, 36, 201, 118]
t3 = [34, 31, 18, 18, 67, 57, 62, 7, 22, 34]
t4 = [90, 10, 60, 186, 61, 49, 14, 24, 56, 20, 79, 84, 44, 59, 29,
118, 25, 156, 310, 76, 26, 44, 23, 62]
t5 = [130, 208, 70, 101, 208]
t6 = [74, 57, 48, 29, 502, 12, 70, 21, 29, 386, 59, 27]
t7 = [55, 320, 56, 104, 220, 239, 47, 246, 176, 182, 33]
t8 = [23, 261, 87, 7, 120, 14, 62, 47, 225, 71, 246, 21, 42, 20, 5,
12, 120, 11, 3, 14, 71, 11, 14, 11, 16, 90, 1, 16, 52, 95]
t9 = [97, 51, 11, 4, 141, 18, 142, 68, 77, 80, 1, 16, 106, 206, 82,
54, 31, 216, 46, 111, 39, 63, 18, 191, 18, 163, 24]
t10 = [50, 44, 102, 72, 22, 39, 3, 15, 197, 188, 79, 88, 46, 5, 5, 36,
22, 139, 210, 97, 30, 23, 13, 14]
t11 = [359, 9, 12, 270, 603, 3, 104, 2, 438]
t12 = [50, 254, 5, 283, 35, 12]
t13 = [487, 18, 100, 7, 98, 5, 85, 91, 43, 230, 3, 130]
t14 = [102, 209, 14, 57, 54, 32, 67, 59, 134, 152, 27, 14, 230, 66,
61, 34]
Tk, tm, p = stats.anderson_ksamp((t1, t2, t3, t4, t5, t6, t7, t8,
t9, t10, t11, t12, t13, t14),
midrank=True)
assert_almost_equal(Tk, 3.294, 3)
assert_array_almost_equal([0.5990, 1.3269, 1.8052, 2.2486, 2.8009],
tm[0:5], 4)
assert_allclose(p, 0.0041, atol=0.00025)
def test_R_kSamples(self):
# test values generates with R package kSamples
# package version 1.2-6 (2017-06-14)
# r1 = 1:100
# continuous case (no ties) --> version 1
# res <- kSamples::ad.test(r1, r1 + 40.5)
# res$ad[1, "T.AD"] # 41.105
# res$ad[1, " asympt. P-value"] # 5.8399e-18
#
# discrete case (ties allowed) --> version 2 (here: midrank=True)
# res$ad[2, "T.AD"] # 41.235
#
# res <- kSamples::ad.test(r1, r1 + .5)
# res$ad[1, "T.AD"] # -1.2824
# res$ad[1, " asympt. P-value"] # 1
# res$ad[2, "T.AD"] # -1.2944
#
# res <- kSamples::ad.test(r1, r1 + 7.5)
# res$ad[1, "T.AD"] # 1.4923
# res$ad[1, " asympt. P-value"] # 0.077501
#
# res <- kSamples::ad.test(r1, r1 + 6)
# res$ad[2, "T.AD"] # 0.63892
# res$ad[2, " asympt. P-value"] # 0.17981
#
# res <- kSamples::ad.test(r1, r1 + 11.5)
# res$ad[1, "T.AD"] # 4.5042
# res$ad[1, " asympt. P-value"] # 0.00545
#
# res <- kSamples::ad.test(r1, r1 + 13.5)
# res$ad[1, "T.AD"] # 6.2982
# res$ad[1, " asympt. P-value"] # 0.00118
x1 = np.linspace(1, 100, 100)
# test case: different distributions;p-value floored at 0.001
# test case for issue #5493 / #8536
with suppress_warnings() as sup:
sup.filter(UserWarning, message='p-value floored')
s, _, p = stats.anderson_ksamp([x1, x1 + 40.5], midrank=False)
assert_almost_equal(s, 41.105, 3)
assert_equal(p, 0.001)
with suppress_warnings() as sup:
sup.filter(UserWarning, message='p-value floored')
s, _, p = stats.anderson_ksamp([x1, x1 + 40.5])
assert_almost_equal(s, 41.235, 3)
assert_equal(p, 0.001)
# test case: similar distributions --> p-value capped at 0.25
with suppress_warnings() as sup:
sup.filter(UserWarning, message='p-value capped')
s, _, p = stats.anderson_ksamp([x1, x1 + .5], midrank=False)
assert_almost_equal(s, -1.2824, 4)
assert_equal(p, 0.25)
with suppress_warnings() as sup:
sup.filter(UserWarning, message='p-value capped')
s, _, p = stats.anderson_ksamp([x1, x1 + .5])
assert_almost_equal(s, -1.2944, 4)
assert_equal(p, 0.25)
# test case: check interpolated p-value in [0.01, 0.25] (no ties)
s, _, p = stats.anderson_ksamp([x1, x1 + 7.5], midrank=False)
assert_almost_equal(s, 1.4923, 4)
assert_allclose(p, 0.0775, atol=0.005, rtol=0)
# test case: check interpolated p-value in [0.01, 0.25] (w/ ties)
s, _, p = stats.anderson_ksamp([x1, x1 + 6])
assert_almost_equal(s, 0.6389, 4)
assert_allclose(p, 0.1798, atol=0.005, rtol=0)
# test extended critical values for p=0.001 and p=0.005
s, _, p = stats.anderson_ksamp([x1, x1 + 11.5], midrank=False)
assert_almost_equal(s, 4.5042, 4)
assert_allclose(p, 0.00545, atol=0.0005, rtol=0)
s, _, p = stats.anderson_ksamp([x1, x1 + 13.5], midrank=False)
assert_almost_equal(s, 6.2982, 4)
assert_allclose(p, 0.00118, atol=0.0001, rtol=0)
def test_not_enough_samples(self):
assert_raises(ValueError, stats.anderson_ksamp, np.ones(5))
def test_no_distinct_observations(self):
assert_raises(ValueError, stats.anderson_ksamp,
(np.ones(5), np.ones(5)))
def test_empty_sample(self):
assert_raises(ValueError, stats.anderson_ksamp, (np.ones(5), []))
def test_result_attributes(self):
# Pass a mixture of lists and arrays
t1 = [38.7, 41.5, 43.8, 44.5, 45.5, 46.0, 47.7, 58.0]
t2 = np.array([39.2, 39.3, 39.7, 41.4, 41.8, 42.9, 43.3, 45.8])
res = stats.anderson_ksamp((t1, t2), midrank=False)
attributes = ('statistic', 'critical_values', 'significance_level')
check_named_results(res, attributes)
class TestAnsari(object):
def test_small(self):
x = [1, 2, 3, 3, 4]
y = [3, 2, 6, 1, 6, 1, 4, 1]
with suppress_warnings() as sup:
sup.filter(UserWarning, "Ties preclude use of exact statistic.")
W, pval = stats.ansari(x, y)
assert_almost_equal(W, 23.5, 11)
assert_almost_equal(pval, 0.13499256881897437, 11)
def test_approx(self):
ramsay = np.array((111, 107, 100, 99, 102, 106, 109, 108, 104, 99,
101, 96, 97, 102, 107, 113, 116, 113, 110, 98))
parekh = np.array((107, 108, 106, 98, 105, 103, 110, 105, 104,
100, 96, 108, 103, 104, 114, 114, 113, 108,
106, 99))
with suppress_warnings() as sup:
sup.filter(UserWarning, "Ties preclude use of exact statistic.")
W, pval = stats.ansari(ramsay, parekh)
assert_almost_equal(W, 185.5, 11)
assert_almost_equal(pval, 0.18145819972867083, 11)
def test_exact(self):
W, pval = stats.ansari([1, 2, 3, 4], [15, 5, 20, 8, 10, 12])
assert_almost_equal(W, 10.0, 11)
assert_almost_equal(pval, 0.533333333333333333, 7)
def test_bad_arg(self):
assert_raises(ValueError, stats.ansari, [], [1])
assert_raises(ValueError, stats.ansari, [1], [])
def test_result_attributes(self):
x = [1, 2, 3, 3, 4]
y = [3, 2, 6, 1, 6, 1, 4, 1]
with suppress_warnings() as sup:
sup.filter(UserWarning, "Ties preclude use of exact statistic.")
res = stats.ansari(x, y)
attributes = ('statistic', 'pvalue')
check_named_results(res, attributes)
class TestBartlett(object):
def test_data(self):
# https://www.itl.nist.gov/div898/handbook/eda/section3/eda357.htm
args = [g1, g2, g3, g4, g5, g6, g7, g8, g9, g10]
T, pval = stats.bartlett(*args)
assert_almost_equal(T, 20.78587342806484, 7)
assert_almost_equal(pval, 0.0136358632781, 7)
def test_bad_arg(self):
# Too few args raises ValueError.
assert_raises(ValueError, stats.bartlett, [1])
def test_result_attributes(self):
args = [g1, g2, g3, g4, g5, g6, g7, g8, g9, g10]
res = stats.bartlett(*args)
attributes = ('statistic', 'pvalue')
check_named_results(res, attributes)
def test_empty_arg(self):
args = (g1, g2, g3, g4, g5, g6, g7, g8, g9, g10, [])
assert_equal((np.nan, np.nan), stats.bartlett(*args))
# temporary fix for issue #9252: only accept 1d input
def test_1d_input(self):
x = np.array([[1, 2], [3, 4]])
assert_raises(ValueError, stats.bartlett, g1, x)
class TestLevene(object):
def test_data(self):
# https://www.itl.nist.gov/div898/handbook/eda/section3/eda35a.htm
args = [g1, g2, g3, g4, g5, g6, g7, g8, g9, g10]
W, pval = stats.levene(*args)
assert_almost_equal(W, 1.7059176930008939, 7)
assert_almost_equal(pval, 0.0990829755522, 7)
def test_trimmed1(self):
# Test that center='trimmed' gives the same result as center='mean'
# when proportiontocut=0.
W1, pval1 = stats.levene(g1, g2, g3, center='mean')
W2, pval2 = stats.levene(g1, g2, g3, center='trimmed',
proportiontocut=0.0)
assert_almost_equal(W1, W2)
assert_almost_equal(pval1, pval2)
def test_trimmed2(self):
x = [1.2, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 100.0]
y = [0.0, 3.0, 3.5, 4.0, 4.5, 5.0, 5.5, 200.0]
np.random.seed(1234)
x2 = np.random.permutation(x)
# Use center='trimmed'
W0, pval0 = stats.levene(x, y, center='trimmed',
proportiontocut=0.125)
W1, pval1 = stats.levene(x2, y, center='trimmed',
proportiontocut=0.125)
# Trim the data here, and use center='mean'
W2, pval2 = stats.levene(x[1:-1], y[1:-1], center='mean')
# Result should be the same.
assert_almost_equal(W0, W2)
assert_almost_equal(W1, W2)
assert_almost_equal(pval1, pval2)
def test_equal_mean_median(self):
x = np.linspace(-1, 1, 21)
np.random.seed(1234)
x2 = np.random.permutation(x)
y = x**3
W1, pval1 = stats.levene(x, y, center='mean')
W2, pval2 = stats.levene(x2, y, center='median')
assert_almost_equal(W1, W2)
assert_almost_equal(pval1, pval2)
def test_bad_keyword(self):
x = np.linspace(-1, 1, 21)
assert_raises(TypeError, stats.levene, x, x, portiontocut=0.1)
def test_bad_center_value(self):
x = np.linspace(-1, 1, 21)
assert_raises(ValueError, stats.levene, x, x, center='trim')
def test_too_few_args(self):
assert_raises(ValueError, stats.levene, [1])
def test_result_attributes(self):
args = [g1, g2, g3, g4, g5, g6, g7, g8, g9, g10]
res = stats.levene(*args)
attributes = ('statistic', 'pvalue')
check_named_results(res, attributes)
# temporary fix for issue #9252: only accept 1d input
def test_1d_input(self):
x = np.array([[1, 2], [3, 4]])
assert_raises(ValueError, stats.levene, g1, x)
class TestBinomP(object):
def test_data(self):
pval = stats.binom_test(100, 250)
assert_almost_equal(pval, 0.0018833009350757682, 11)
pval = stats.binom_test(201, 405)
assert_almost_equal(pval, 0.92085205962670713, 11)
pval = stats.binom_test([682, 243], p=3.0/4)
assert_almost_equal(pval, 0.38249155957481695, 11)
def test_bad_len_x(self):
# Length of x must be 1 or 2.
assert_raises(ValueError, stats.binom_test, [1, 2, 3])
def test_bad_n(self):
# len(x) is 1, but n is invalid.
# Missing n
assert_raises(ValueError, stats.binom_test, [100])
# n less than x[0]
assert_raises(ValueError, stats.binom_test, [100], n=50)
def test_bad_p(self):
assert_raises(ValueError, stats.binom_test, [50, 50], p=2.0)
def test_alternatives(self):
res = stats.binom_test(51, 235, p=1./6, alternative='less')
assert_almost_equal(res, 0.982022657605858)
res = stats.binom_test(51, 235, p=1./6, alternative='greater')
assert_almost_equal(res, 0.02654424571169085)
res = stats.binom_test(51, 235, p=1./6, alternative='two-sided')
assert_almost_equal(res, 0.0437479701823997)
class TestFligner(object):
def test_data(self):
# numbers from R: fligner.test in package stats
x1 = np.arange(5)
assert_array_almost_equal(stats.fligner(x1, x1**2),
(3.2282229927203536, 0.072379187848207877),
11)
def test_trimmed1(self):
# Perturb input to break ties in the transformed data
# See https://github.com/scipy/scipy/pull/8042 for more details
rs = np.random.RandomState(123)
_perturb = lambda g: (np.asarray(g) + 1e-10*rs.randn(len(g))).tolist()
g1_ = _perturb(g1)
g2_ = _perturb(g2)
g3_ = _perturb(g3)
# Test that center='trimmed' gives the same result as center='mean'
# when proportiontocut=0.
Xsq1, pval1 = stats.fligner(g1_, g2_, g3_, center='mean')
Xsq2, pval2 = stats.fligner(g1_, g2_, g3_, center='trimmed',
proportiontocut=0.0)
assert_almost_equal(Xsq1, Xsq2)
assert_almost_equal(pval1, pval2)
def test_trimmed2(self):
x = [1.2, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 100.0]
y = [0.0, 3.0, 3.5, 4.0, 4.5, 5.0, 5.5, 200.0]
# Use center='trimmed'
Xsq1, pval1 = stats.fligner(x, y, center='trimmed',
proportiontocut=0.125)
# Trim the data here, and use center='mean'
Xsq2, pval2 = stats.fligner(x[1:-1], y[1:-1], center='mean')
# Result should be the same.
assert_almost_equal(Xsq1, Xsq2)
assert_almost_equal(pval1, pval2)
# The following test looks reasonable at first, but fligner() uses the
# function stats.rankdata(), and in one of the cases in this test,
# there are ties, while in the other (because of normal rounding
# errors) there are not. This difference leads to differences in the
# third significant digit of W.
#
#def test_equal_mean_median(self):
# x = np.linspace(-1,1,21)
# y = x**3
# W1, pval1 = stats.fligner(x, y, center='mean')
# W2, pval2 = stats.fligner(x, y, center='median')
# assert_almost_equal(W1, W2)
# assert_almost_equal(pval1, pval2)
def test_bad_keyword(self):
x = np.linspace(-1, 1, 21)
assert_raises(TypeError, stats.fligner, x, x, portiontocut=0.1)
def test_bad_center_value(self):
x = np.linspace(-1, 1, 21)
assert_raises(ValueError, stats.fligner, x, x, center='trim')
def test_bad_num_args(self):
# Too few args raises ValueError.
assert_raises(ValueError, stats.fligner, [1])
def test_empty_arg(self):
x = np.arange(5)
assert_equal((np.nan, np.nan), stats.fligner(x, x**2, []))
class TestMood(object):
def test_mood(self):
# numbers from R: mood.test in package stats
x1 = np.arange(5)
assert_array_almost_equal(stats.mood(x1, x1**2),
(-1.3830857299399906, 0.16663858066771478),
11)
def test_mood_order_of_args(self):
# z should change sign when the order of arguments changes, pvalue
# should not change
np.random.seed(1234)
x1 = np.random.randn(10, 1)
x2 = np.random.randn(15, 1)
z1, p1 = stats.mood(x1, x2)
z2, p2 = stats.mood(x2, x1)
assert_array_almost_equal([z1, p1], [-z2, p2])
def test_mood_with_axis_none(self):
# Test with axis = None, compare with results from R
x1 = [-0.626453810742332, 0.183643324222082, -0.835628612410047,
1.59528080213779, 0.329507771815361, -0.820468384118015,
0.487429052428485, 0.738324705129217, 0.575781351653492,
-0.305388387156356, 1.51178116845085, 0.389843236411431,
-0.621240580541804, -2.2146998871775, 1.12493091814311,
-0.0449336090152309, -0.0161902630989461, 0.943836210685299,
0.821221195098089, 0.593901321217509]
x2 = [-0.896914546624981, 0.184849184646742, 1.58784533120882,
-1.13037567424629, -0.0802517565509893, 0.132420284381094,
0.707954729271733, -0.23969802417184, 1.98447393665293,
-0.138787012119665, 0.417650750792556, 0.981752777463662,
-0.392695355503813, -1.03966897694891, 1.78222896030858,
-2.31106908460517, 0.878604580921265, 0.035806718015226,
1.01282869212708, 0.432265154539617, 2.09081920524915,
-1.19992581964387, 1.58963820029007, 1.95465164222325,
0.00493777682814261, -2.45170638784613, 0.477237302613617,
-0.596558168631403, 0.792203270299649, 0.289636710177348]
x1 = np.array(x1)
x2 = np.array(x2)
x1.shape = (10, 2)
x2.shape = (15, 2)
assert_array_almost_equal(stats.mood(x1, x2, axis=None),
[-1.31716607555, 0.18778296257])
def test_mood_2d(self):
# Test if the results of mood test in 2-D case are consistent with the
# R result for the same inputs. Numbers from R mood.test().
ny = 5
np.random.seed(1234)
x1 = np.random.randn(10, ny)
x2 = np.random.randn(15, ny)
z_vectest, pval_vectest = stats.mood(x1, x2)
for j in range(ny):
assert_array_almost_equal([z_vectest[j], pval_vectest[j]],
stats.mood(x1[:, j], x2[:, j]))
# inverse order of dimensions
x1 = x1.transpose()
x2 = x2.transpose()
z_vectest, pval_vectest = stats.mood(x1, x2, axis=1)
for i in range(ny):
# check axis handling is self consistent
assert_array_almost_equal([z_vectest[i], pval_vectest[i]],
stats.mood(x1[i, :], x2[i, :]))
def test_mood_3d(self):
shape = (10, 5, 6)
np.random.seed(1234)
x1 = np.random.randn(*shape)
x2 = np.random.randn(*shape)
for axis in range(3):
z_vectest, pval_vectest = stats.mood(x1, x2, axis=axis)
# Tests that result for 3-D arrays is equal to that for the
# same calculation on a set of 1-D arrays taken from the
# 3-D array
axes_idx = ([1, 2], [0, 2], [0, 1]) # the two axes != axis
for i in range(shape[axes_idx[axis][0]]):
for j in range(shape[axes_idx[axis][1]]):
if axis == 0:
slice1 = x1[:, i, j]
slice2 = x2[:, i, j]
elif axis == 1:
slice1 = x1[i, :, j]
slice2 = x2[i, :, j]
else:
slice1 = x1[i, j, :]
slice2 = x2[i, j, :]
assert_array_almost_equal([z_vectest[i, j],
pval_vectest[i, j]],
stats.mood(slice1, slice2))
def test_mood_bad_arg(self):
# Raise ValueError when the sum of the lengths of the args is
# less than 3
assert_raises(ValueError, stats.mood, [1], [])
class TestProbplot(object):
def test_basic(self):
np.random.seed(12345)
x = stats.norm.rvs(size=20)
osm, osr = stats.probplot(x, fit=False)
osm_expected = [-1.8241636, -1.38768012, -1.11829229, -0.91222575,
-0.73908135, -0.5857176, -0.44506467, -0.31273668,
-0.18568928, -0.06158146, 0.06158146, 0.18568928,
0.31273668, 0.44506467, 0.5857176, 0.73908135,
0.91222575, 1.11829229, 1.38768012, 1.8241636]
assert_allclose(osr, np.sort(x))
assert_allclose(osm, osm_expected)
res, res_fit = stats.probplot(x, fit=True)
res_fit_expected = [1.05361841, 0.31297795, 0.98741609]
assert_allclose(res_fit, res_fit_expected)
def test_sparams_keyword(self):
np.random.seed(123456)
x = stats.norm.rvs(size=100)
# Check that None, () and 0 (loc=0, for normal distribution) all work
# and give the same results
osm1, osr1 = stats.probplot(x, sparams=None, fit=False)
osm2, osr2 = stats.probplot(x, sparams=0, fit=False)
osm3, osr3 = stats.probplot(x, sparams=(), fit=False)
assert_allclose(osm1, osm2)
assert_allclose(osm1, osm3)
assert_allclose(osr1, osr2)
assert_allclose(osr1, osr3)
# Check giving (loc, scale) params for normal distribution
osm, osr = stats.probplot(x, sparams=(), fit=False)
def test_dist_keyword(self):
np.random.seed(12345)
x = stats.norm.rvs(size=20)
osm1, osr1 = stats.probplot(x, fit=False, dist='t', sparams=(3,))
osm2, osr2 = stats.probplot(x, fit=False, dist=stats.t, sparams=(3,))
assert_allclose(osm1, osm2)
assert_allclose(osr1, osr2)
assert_raises(ValueError, stats.probplot, x, dist='wrong-dist-name')
assert_raises(AttributeError, stats.probplot, x, dist=[])
class custom_dist(object):
"""Some class that looks just enough like a distribution."""
def ppf(self, q):
return stats.norm.ppf(q, loc=2)
osm1, osr1 = stats.probplot(x, sparams=(2,), fit=False)
osm2, osr2 = stats.probplot(x, dist=custom_dist(), fit=False)
assert_allclose(osm1, osm2)
assert_allclose(osr1, osr2)
@pytest.mark.skipif(not have_matplotlib, reason="no matplotlib")
def test_plot_kwarg(self):
np.random.seed(7654321)
fig = plt.figure()
fig.add_subplot(111)
x = stats.t.rvs(3, size=100)
res1, fitres1 = stats.probplot(x, plot=plt)
plt.close()
res2, fitres2 = stats.probplot(x, plot=None)
res3 = stats.probplot(x, fit=False, plot=plt)
plt.close()
res4 = stats.probplot(x, fit=False, plot=None)
# Check that results are consistent between combinations of `fit` and
# `plot` keywords.
assert_(len(res1) == len(res2) == len(res3) == len(res4) == 2)
assert_allclose(res1, res2)
assert_allclose(res1, res3)
assert_allclose(res1, res4)
assert_allclose(fitres1, fitres2)
# Check that a Matplotlib Axes object is accepted
fig = plt.figure()
ax = fig.add_subplot(111)
stats.probplot(x, fit=False, plot=ax)
plt.close()
def test_probplot_bad_args(self):
# Raise ValueError when given an invalid distribution.
assert_raises(ValueError, stats.probplot, [1], dist="plate_of_shrimp")
def test_empty(self):
assert_equal(stats.probplot([], fit=False),
(np.array([]), np.array([])))
assert_equal(stats.probplot([], fit=True),
((np.array([]), np.array([])),
(np.nan, np.nan, 0.0)))
def test_array_of_size_one(self):
with np.errstate(invalid='ignore'):
assert_equal(stats.probplot([1], fit=True),
((np.array([0.]), np.array([1])),
(np.nan, np.nan, 0.0)))
class TestWilcoxon(object):
def test_wilcoxon_bad_arg(self):
# Raise ValueError when two args of different lengths are given or
# zero_method is unknown.
assert_raises(ValueError, stats.wilcoxon, [1], [1, 2])
assert_raises(ValueError, stats.wilcoxon, [1, 2], [1, 2], "dummy")
assert_raises(ValueError, stats.wilcoxon, [1, 2], [1, 2],
alternative="dummy")
def test_zero_diff(self):
x = np.arange(20)
# pratt and wilcox do not work if x - y == 0
assert_raises(ValueError, stats.wilcoxon, x, x, "wilcox")
assert_raises(ValueError, stats.wilcoxon, x, x, "pratt")
# ranksum is n*(n+1)/2, split in half if method == "zsplit"
assert_equal(stats.wilcoxon(x, x, "zsplit"), (20*21/4, 1.0))
def test_pratt(self):
# regression test for gh-6805: p-value matches value from R package
# coin (wilcoxsign_test) reported in the issue
x = [1, 2, 3, 4]
y = [1, 2, 3, 5]
with suppress_warnings() as sup:
sup.filter(UserWarning, message="Sample size too small")
res = stats.wilcoxon(x, y, zero_method="pratt")
assert_allclose(res, (0.0, 0.31731050786291415))
def test_wilcoxon_arg_type(self):
# Should be able to accept list as arguments.
# Address issue 6070.
arr = [1, 2, 3, 0, -1, 3, 1, 2, 1, 1, 2]
_ = stats.wilcoxon(arr, zero_method="pratt")
_ = stats.wilcoxon(arr, zero_method="zsplit")
_ = stats.wilcoxon(arr, zero_method="wilcox")
def test_accuracy_wilcoxon(self):
freq = [1, 4, 16, 15, 8, 4, 5, 1, 2]
nums = range(-4, 5)
x = np.concatenate([[u] * v for u, v in zip(nums, freq)])
y = np.zeros(x.size)
T, p = stats.wilcoxon(x, y, "pratt")
assert_allclose(T, 423)
assert_allclose(p, 0.0031724568006762576)
T, p = stats.wilcoxon(x, y, "zsplit")
assert_allclose(T, 441)
assert_allclose(p, 0.0032145343172473055)
T, p = stats.wilcoxon(x, y, "wilcox")
assert_allclose(T, 327)
assert_allclose(p, 0.00641346115861)
# Test the 'correction' option, using values computed in R with:
# > wilcox.test(x, y, paired=TRUE, exact=FALSE, correct={FALSE,TRUE})
x = np.array([120, 114, 181, 188, 180, 146, 121, 191, 132, 113, 127, 112])
y = np.array([133, 143, 119, 189, 112, 199, 198, 113, 115, 121, 142, 187])
T, p = stats.wilcoxon(x, y, correction=False)
assert_equal(T, 34)
assert_allclose(p, 0.6948866, rtol=1e-6)
T, p = stats.wilcoxon(x, y, correction=True)
assert_equal(T, 34)
assert_allclose(p, 0.7240817, rtol=1e-6)
def test_wilcoxon_result_attributes(self):
x = np.array([120, 114, 181, 188, 180, 146, 121, 191, 132, 113, 127, 112])
y = np.array([133, 143, 119, 189, 112, 199, 198, 113, 115, 121, 142, 187])
res = stats.wilcoxon(x, y, correction=False)
attributes = ('statistic', 'pvalue')
check_named_results(res, attributes)
def test_wilcoxon_tie(self):
# Regression test for gh-2391.
# Corresponding R code is:
# > result = wilcox.test(rep(0.1, 10), exact=FALSE, correct=FALSE)
# > result$p.value
# [1] 0.001565402
# > result = wilcox.test(rep(0.1, 10), exact=FALSE, correct=TRUE)
# > result$p.value
# [1] 0.001904195
stat, p = stats.wilcoxon([0.1] * 10)
expected_p = 0.001565402
assert_equal(stat, 0)
assert_allclose(p, expected_p, rtol=1e-6)
stat, p = stats.wilcoxon([0.1] * 10, correction=True)
expected_p = 0.001904195
assert_equal(stat, 0)
assert_allclose(p, expected_p, rtol=1e-6)
def test_onesided(self):
# tested against "R version 3.4.1 (2017-06-30)"
# x <- c(125, 115, 130, 140, 140, 115, 140, 125, 140, 135)
# y <- c(110, 122, 125, 120, 140, 124, 123, 137, 135, 145)
# cfg <- list(x = x, y = y, paired = TRUE, exact = FALSE)
# do.call(wilcox.test, c(cfg, list(alternative = "less", correct = FALSE)))
# do.call(wilcox.test, c(cfg, list(alternative = "less", correct = TRUE)))
# do.call(wilcox.test, c(cfg, list(alternative = "greater", correct = FALSE)))
# do.call(wilcox.test, c(cfg, list(alternative = "greater", correct = TRUE)))
x = [125, 115, 130, 140, 140, 115, 140, 125, 140, 135]
y = [110, 122, 125, 120, 140, 124, 123, 137, 135, 145]
with suppress_warnings() as sup:
sup.filter(UserWarning, message="Sample size too small")
w, p = stats.wilcoxon(x, y, alternative="less")
assert_equal(w, 27)
assert_almost_equal(p, 0.7031847, decimal=6)
with suppress_warnings() as sup:
sup.filter(UserWarning, message="Sample size too small")
w, p = stats.wilcoxon(x, y, alternative="less", correction=True)
assert_equal(w, 27)
assert_almost_equal(p, 0.7233656, decimal=6)
with suppress_warnings() as sup:
sup.filter(UserWarning, message="Sample size too small")
w, p = stats.wilcoxon(x, y, alternative="greater")
assert_equal(w, 27)
assert_almost_equal(p, 0.2968153, decimal=6)
with suppress_warnings() as sup:
sup.filter(UserWarning, message="Sample size too small")
w, p = stats.wilcoxon(x, y, alternative="greater", correction=True)
assert_equal(w, 27)
assert_almost_equal(p, 0.3176447, decimal=6)
class TestKstat(object):
def test_moments_normal_distribution(self):
np.random.seed(32149)
data = np.random.randn(12345)
moments = [stats.kstat(data, n) for n in [1, 2, 3, 4]]
expected = [0.011315, 1.017931, 0.05811052, 0.0754134]
assert_allclose(moments, expected, rtol=1e-4)
# test equivalence with `stats.moment`
m1 = stats.moment(data, moment=1)
m2 = stats.moment(data, moment=2)
m3 = stats.moment(data, moment=3)
assert_allclose((m1, m2, m3), expected[:-1], atol=0.02, rtol=1e-2)
def test_empty_input(self):
assert_raises(ValueError, stats.kstat, [])
def test_nan_input(self):
data = np.arange(10.)
data[6] = np.nan
assert_equal(stats.kstat(data), np.nan)
def test_kstat_bad_arg(self):
# Raise ValueError if n > 4 or n < 1.
data = np.arange(10)
for n in [0, 4.001]:
assert_raises(ValueError, stats.kstat, data, n=n)
class TestKstatVar(object):
def test_empty_input(self):
assert_raises(ValueError, stats.kstatvar, [])
def test_nan_input(self):
data = np.arange(10.)
data[6] = np.nan
assert_equal(stats.kstat(data), np.nan)
def test_bad_arg(self):
# Raise ValueError is n is not 1 or 2.
data = [1]
n = 10
assert_raises(ValueError, stats.kstatvar, data, n=n)
class TestPpccPlot(object):
def setup_method(self):
np.random.seed(7654321)
self.x = stats.loggamma.rvs(5, size=500) + 5
def test_basic(self):
N = 5
svals, ppcc = stats.ppcc_plot(self.x, -10, 10, N=N)
ppcc_expected = [0.21139644, 0.21384059, 0.98766719, 0.97980182,
0.93519298]
assert_allclose(svals, np.linspace(-10, 10, num=N))
assert_allclose(ppcc, ppcc_expected)
def test_dist(self):
# Test that we can specify distributions both by name and as objects.
svals1, ppcc1 = stats.ppcc_plot(self.x, -10, 10, dist='tukeylambda')
svals2, ppcc2 = stats.ppcc_plot(self.x, -10, 10,
dist=stats.tukeylambda)
assert_allclose(svals1, svals2, rtol=1e-20)
assert_allclose(ppcc1, ppcc2, rtol=1e-20)
# Test that 'tukeylambda' is the default dist
svals3, ppcc3 = stats.ppcc_plot(self.x, -10, 10)
assert_allclose(svals1, svals3, rtol=1e-20)
assert_allclose(ppcc1, ppcc3, rtol=1e-20)
@pytest.mark.skipif(not have_matplotlib, reason="no matplotlib")
def test_plot_kwarg(self):
# Check with the matplotlib.pyplot module
fig = plt.figure()
ax = fig.add_subplot(111)
stats.ppcc_plot(self.x, -20, 20, plot=plt)
fig.delaxes(ax)
# Check that a Matplotlib Axes object is accepted
ax = fig.add_subplot(111)
stats.ppcc_plot(self.x, -20, 20, plot=ax)
plt.close()
def test_invalid_inputs(self):
# `b` has to be larger than `a`
assert_raises(ValueError, stats.ppcc_plot, self.x, 1, 0)
# Raise ValueError when given an invalid distribution.
assert_raises(ValueError, stats.ppcc_plot, [1, 2, 3], 0, 1,
dist="plate_of_shrimp")
def test_empty(self):
# For consistency with probplot return for one empty array,
# ppcc contains all zeros and svals is the same as for normal array
# input.
svals, ppcc = stats.ppcc_plot([], 0, 1)
assert_allclose(svals, np.linspace(0, 1, num=80))
assert_allclose(ppcc, np.zeros(80, dtype=float))
class TestPpccMax(object):
def test_ppcc_max_bad_arg(self):
# Raise ValueError when given an invalid distribution.
data = [1]
assert_raises(ValueError, stats.ppcc_max, data, dist="plate_of_shrimp")
def test_ppcc_max_basic(self):
np.random.seed(1234567)
x = stats.tukeylambda.rvs(-0.7, loc=2, scale=0.5, size=10000) + 1e4
# On Python 2.6 the result is accurate to 5 decimals. On Python >= 2.7
# it is accurate up to 16 decimals
assert_almost_equal(stats.ppcc_max(x), -0.71215366521264145, decimal=5)
def test_dist(self):
np.random.seed(1234567)
x = stats.tukeylambda.rvs(-0.7, loc=2, scale=0.5, size=10000) + 1e4
# Test that we can specify distributions both by name and as objects.
max1 = stats.ppcc_max(x, dist='tukeylambda')
max2 = stats.ppcc_max(x, dist=stats.tukeylambda)
assert_almost_equal(max1, -0.71215366521264145, decimal=5)
assert_almost_equal(max2, -0.71215366521264145, decimal=5)
# Test that 'tukeylambda' is the default dist
max3 = stats.ppcc_max(x)
assert_almost_equal(max3, -0.71215366521264145, decimal=5)
def test_brack(self):
np.random.seed(1234567)
x = stats.tukeylambda.rvs(-0.7, loc=2, scale=0.5, size=10000) + 1e4
assert_raises(ValueError, stats.ppcc_max, x, brack=(0.0, 1.0, 0.5))
# On Python 2.6 the result is accurate to 5 decimals. On Python >= 2.7
# it is accurate up to 16 decimals
assert_almost_equal(stats.ppcc_max(x, brack=(0, 1)),
-0.71215366521264145, decimal=5)
# On Python 2.6 the result is accurate to 5 decimals. On Python >= 2.7
# it is accurate up to 16 decimals
assert_almost_equal(stats.ppcc_max(x, brack=(-2, 2)),
-0.71215366521264145, decimal=5)
class TestBoxcox_llf(object):
def test_basic(self):
np.random.seed(54321)
x = stats.norm.rvs(size=10000, loc=10)
lmbda = 1
llf = stats.boxcox_llf(lmbda, x)
llf_expected = -x.size / 2. * np.log(np.sum(x.std()**2))
assert_allclose(llf, llf_expected)
def test_array_like(self):
np.random.seed(54321)
x = stats.norm.rvs(size=100, loc=10)
lmbda = 1
llf = stats.boxcox_llf(lmbda, x)
llf2 = stats.boxcox_llf(lmbda, list(x))
assert_allclose(llf, llf2, rtol=1e-12)
def test_2d_input(self):
# Note: boxcox_llf() was already working with 2-D input (sort of), so
# keep it like that. boxcox() doesn't work with 2-D input though, due
# to brent() returning a scalar.
np.random.seed(54321)
x = stats.norm.rvs(size=100, loc=10)
lmbda = 1
llf = stats.boxcox_llf(lmbda, x)
llf2 = stats.boxcox_llf(lmbda, np.vstack([x, x]).T)
assert_allclose([llf, llf], llf2, rtol=1e-12)
def test_empty(self):
assert_(np.isnan(stats.boxcox_llf(1, [])))
def test_gh_6873(self):
# Regression test for gh-6873.
# This example was taken from gh-7534, a duplicate of gh-6873.
data = [198.0, 233.0, 233.0, 392.0]
llf = stats.boxcox_llf(-8, data)
# The expected value was computed with mpmath.
assert_allclose(llf, -17.93934208579061)
# This is the data from github user Qukaiyi, given as an example
# of a data set that caused boxcox to fail.
_boxcox_data = [
15957, 112079, 1039553, 711775, 173111, 307382, 183155, 53366, 760875,
207500, 160045, 473714, 40194, 440319, 133261, 265444, 155590, 36660,
904939, 55108, 138391, 339146, 458053, 63324, 1377727, 1342632, 41575,
68685, 172755, 63323, 368161, 199695, 538214, 167760, 388610, 398855,
1001873, 364591, 1320518, 194060, 194324, 2318551, 196114, 64225, 272000,
198668, 123585, 86420, 1925556, 695798, 88664, 46199, 759135, 28051,
345094, 1977752, 51778, 82746, 638126, 2560910, 45830, 140576, 1603787,
57371, 548730, 5343629, 2298913, 998813, 2156812, 423966, 68350, 145237,
131935, 1600305, 342359, 111398, 1409144, 281007, 60314, 242004, 113418,
246211, 61940, 95858, 957805, 40909, 307955, 174159, 124278, 241193,
872614, 304180, 146719, 64361, 87478, 509360, 167169, 933479, 620561,
483333, 97416, 143518, 286905, 597837, 2556043, 89065, 69944, 196858,
88883, 49379, 916265, 1527392, 626954, 54415, 89013, 2883386, 106096,
402697, 45578, 349852, 140379, 34648, 757343, 1305442, 2054757, 121232,
606048, 101492, 51426, 1820833, 83412, 136349, 1379924, 505977, 1303486,
95853, 146451, 285422, 2205423, 259020, 45864, 684547, 182014, 784334,
174793, 563068, 170745, 1195531, 63337, 71833, 199978, 2330904, 227335,
898280, 75294, 2011361, 116771, 157489, 807147, 1321443, 1148635, 2456524,
81839, 1228251, 97488, 1051892, 75397, 3009923, 2732230, 90923, 39735,
132433, 225033, 337555, 1204092, 686588, 1062402, 40362, 1361829, 1497217,
150074, 551459, 2019128, 39581, 45349, 1117187, 87845, 1877288, 164448,
10338362, 24942, 64737, 769946, 2469124, 2366997, 259124, 2667585, 29175,
56250, 74450, 96697, 5920978, 838375, 225914, 119494, 206004, 430907,
244083, 219495, 322239, 407426, 618748, 2087536, 2242124, 4736149, 124624,
406305, 240921, 2675273, 4425340, 821457, 578467, 28040, 348943, 48795,
145531, 52110, 1645730, 1768364, 348363, 85042, 2673847, 81935, 169075,
367733, 135474, 383327, 1207018, 93481, 5934183, 352190, 636533, 145870,
55659, 146215, 73191, 248681, 376907, 1606620, 169381, 81164, 246390,
236093, 885778, 335969, 49266, 381430, 307437, 350077, 34346, 49340,
84715, 527120, 40163, 46898, 4609439, 617038, 2239574, 159905, 118337,
120357, 430778, 3799158, 3516745, 54198, 2970796, 729239, 97848, 6317375,
887345, 58198, 88111, 867595, 210136, 1572103, 1420760, 574046, 845988,
509743, 397927, 1119016, 189955, 3883644, 291051, 126467, 1239907, 2556229,
411058, 657444, 2025234, 1211368, 93151, 577594, 4842264, 1531713, 305084,
479251, 20591, 1466166, 137417, 897756, 594767, 3606337, 32844, 82426,
1294831, 57174, 290167, 322066, 813146, 5671804, 4425684, 895607, 450598,
1048958, 232844, 56871, 46113, 70366, 701618, 97739, 157113, 865047,
194810, 1501615, 1765727, 38125, 2733376, 40642, 437590, 127337, 106310,
4167579, 665303, 809250, 1210317, 45750, 1853687, 348954, 156786, 90793,
1885504, 281501, 3902273, 359546, 797540, 623508, 3672775, 55330, 648221,
266831, 90030, 7118372, 735521, 1009925, 283901, 806005, 2434897, 94321,
309571, 4213597, 2213280, 120339, 64403, 8155209, 1686948, 4327743,
1868312, 135670, 3189615, 1569446, 706058, 58056, 2438625, 520619, 105201,
141961, 179990, 1351440, 3148662, 2804457, 2760144, 70775, 33807, 1926518,
2362142, 186761, 240941, 97860, 1040429, 1431035, 78892, 484039, 57845,
724126, 3166209, 175913, 159211, 1182095, 86734, 1921472, 513546, 326016,
1891609
]
class TestBoxcox(object):
def test_fixed_lmbda(self):
np.random.seed(12345)
x = stats.loggamma.rvs(5, size=50) + 5
xt = stats.boxcox(x, lmbda=1)
assert_allclose(xt, x - 1)
xt = stats.boxcox(x, lmbda=-1)
assert_allclose(xt, 1 - 1/x)
xt = stats.boxcox(x, lmbda=0)
assert_allclose(xt, np.log(x))
# Also test that array_like input works
xt = stats.boxcox(list(x), lmbda=0)
assert_allclose(xt, np.log(x))
def test_lmbda_None(self):
np.random.seed(1234567)
# Start from normal rv's, do inverse transform to check that
# optimization function gets close to the right answer.
np.random.seed(1245)
lmbda = 2.5
x = stats.norm.rvs(loc=10, size=50000)
x_inv = (x * lmbda + 1)**(-lmbda)
xt, maxlog = stats.boxcox(x_inv)
assert_almost_equal(maxlog, -1 / lmbda, decimal=2)
def test_alpha(self):
np.random.seed(1234)
x = stats.loggamma.rvs(5, size=50) + 5
# Some regular values for alpha, on a small sample size
_, _, interval = stats.boxcox(x, alpha=0.75)
assert_allclose(interval, [4.004485780226041, 5.138756355035744])
_, _, interval = stats.boxcox(x, alpha=0.05)
assert_allclose(interval, [1.2138178554857557, 8.209033272375663])
# Try some extreme values, see we don't hit the N=500 limit
x = stats.loggamma.rvs(7, size=500) + 15
_, _, interval = stats.boxcox(x, alpha=0.001)
assert_allclose(interval, [0.3988867, 11.40553131])
_, _, interval = stats.boxcox(x, alpha=0.999)
assert_allclose(interval, [5.83316246, 5.83735292])
def test_boxcox_bad_arg(self):
# Raise ValueError if any data value is negative.
x = np.array([-1])
assert_raises(ValueError, stats.boxcox, x)
def test_empty(self):
assert_(stats.boxcox([]).shape == (0,))
def test_gh_6873(self):
# Regression test for gh-6873.
y, lam = stats.boxcox(_boxcox_data)
# The expected value of lam was computed with the function
# powerTransform in the R library 'car'. I trust that value
# to only about five significant digits.
assert_allclose(lam, -0.051654, rtol=1e-5)
class TestBoxcoxNormmax(object):
def setup_method(self):
np.random.seed(12345)
self.x = stats.loggamma.rvs(5, size=50) + 5
def test_pearsonr(self):
maxlog = stats.boxcox_normmax(self.x)
assert_allclose(maxlog, 1.804465, rtol=1e-6)
def test_mle(self):
maxlog = stats.boxcox_normmax(self.x, method='mle')
assert_allclose(maxlog, 1.758101, rtol=1e-6)
# Check that boxcox() uses 'mle'
_, maxlog_boxcox = stats.boxcox(self.x)
assert_allclose(maxlog_boxcox, maxlog)
def test_all(self):
maxlog_all = stats.boxcox_normmax(self.x, method='all')
assert_allclose(maxlog_all, [1.804465, 1.758101], rtol=1e-6)
class TestBoxcoxNormplot(object):
def setup_method(self):
np.random.seed(7654321)
self.x = stats.loggamma.rvs(5, size=500) + 5
def test_basic(self):
N = 5
lmbdas, ppcc = stats.boxcox_normplot(self.x, -10, 10, N=N)
ppcc_expected = [0.57783375, 0.83610988, 0.97524311, 0.99756057,
0.95843297]
assert_allclose(lmbdas, np.linspace(-10, 10, num=N))
assert_allclose(ppcc, ppcc_expected)
@pytest.mark.skipif(not have_matplotlib, reason="no matplotlib")
def test_plot_kwarg(self):
# Check with the matplotlib.pyplot module
fig = plt.figure()
ax = fig.add_subplot(111)
stats.boxcox_normplot(self.x, -20, 20, plot=plt)
fig.delaxes(ax)
# Check that a Matplotlib Axes object is accepted
ax = fig.add_subplot(111)
stats.boxcox_normplot(self.x, -20, 20, plot=ax)
plt.close()
def test_invalid_inputs(self):
# `lb` has to be larger than `la`
assert_raises(ValueError, stats.boxcox_normplot, self.x, 1, 0)
# `x` can not contain negative values
assert_raises(ValueError, stats.boxcox_normplot, [-1, 1], 0, 1)
def test_empty(self):
assert_(stats.boxcox_normplot([], 0, 1).size == 0)
class TestYeojohnson_llf(object):
def test_array_like(self):
np.random.seed(54321)
x = stats.norm.rvs(size=100, loc=0)
lmbda = 1
llf = stats.yeojohnson_llf(lmbda, x)
llf2 = stats.yeojohnson_llf(lmbda, list(x))
assert_allclose(llf, llf2, rtol=1e-12)
def test_2d_input(self):
np.random.seed(54321)
x = stats.norm.rvs(size=100, loc=10)
lmbda = 1
llf = stats.yeojohnson_llf(lmbda, x)
llf2 = stats.yeojohnson_llf(lmbda, np.vstack([x, x]).T)
assert_allclose([llf, llf], llf2, rtol=1e-12)
def test_empty(self):
assert_(np.isnan(stats.yeojohnson_llf(1, [])))
class TestYeojohnson(object):
def test_fixed_lmbda(self):
np.random.seed(12345)
# Test positive input
x = stats.loggamma.rvs(5, size=50) + 5
assert np.all(x > 0)
xt = stats.yeojohnson(x, lmbda=1)
assert_allclose(xt, x)
xt = stats.yeojohnson(x, lmbda=-1)
assert_allclose(xt, 1 - 1 / (x + 1))
xt = stats.yeojohnson(x, lmbda=0)
assert_allclose(xt, np.log(x + 1))
xt = stats.yeojohnson(x, lmbda=1)
assert_allclose(xt, x)
# Test negative input
x = stats.loggamma.rvs(5, size=50) - 5
assert np.all(x < 0)
xt = stats.yeojohnson(x, lmbda=2)
assert_allclose(xt, -np.log(-x + 1))
xt = stats.yeojohnson(x, lmbda=1)
assert_allclose(xt, x)
xt = stats.yeojohnson(x, lmbda=3)
assert_allclose(xt, 1 / (-x + 1) - 1)
# test both positive and negative input
x = stats.loggamma.rvs(5, size=50) - 2
assert not np.all(x < 0)
assert not np.all(x >= 0)
pos = x >= 0
xt = stats.yeojohnson(x, lmbda=1)
assert_allclose(xt[pos], x[pos])
xt = stats.yeojohnson(x, lmbda=-1)
assert_allclose(xt[pos], 1 - 1 / (x[pos] + 1))
xt = stats.yeojohnson(x, lmbda=0)
assert_allclose(xt[pos], np.log(x[pos] + 1))
xt = stats.yeojohnson(x, lmbda=1)
assert_allclose(xt[pos], x[pos])
neg = ~pos
xt = stats.yeojohnson(x, lmbda=2)
assert_allclose(xt[neg], -np.log(-x[neg] + 1))
xt = stats.yeojohnson(x, lmbda=1)
assert_allclose(xt[neg], x[neg])
xt = stats.yeojohnson(x, lmbda=3)
assert_allclose(xt[neg], 1 / (-x[neg] + 1) - 1)
@pytest.mark.parametrize('lmbda', [0, .1, .5, 2])
def test_lmbda_None(self, lmbda):
# Start from normal rv's, do inverse transform to check that
# optimization function gets close to the right answer.
def _inverse_transform(x, lmbda):
x_inv = np.zeros(x.shape, dtype=x.dtype)
pos = x >= 0
# when x >= 0
if abs(lmbda) < np.spacing(1.):
x_inv[pos] = np.exp(x[pos]) - 1
else: # lmbda != 0
x_inv[pos] = np.power(x[pos] * lmbda + 1, 1 / lmbda) - 1
# when x < 0
if abs(lmbda - 2) > np.spacing(1.):
x_inv[~pos] = 1 - np.power(-(2 - lmbda) * x[~pos] + 1,
1 / (2 - lmbda))
else: # lmbda == 2
x_inv[~pos] = 1 - np.exp(-x[~pos])
return x_inv
np.random.seed(1234567)
n_samples = 20000
x = np.random.normal(loc=0, scale=1, size=(n_samples))
x_inv = _inverse_transform(x, lmbda)
xt, maxlog = stats.yeojohnson(x_inv)
assert_allclose(maxlog, lmbda, atol=1e-2)
assert_almost_equal(0, np.linalg.norm(x - xt) / n_samples, decimal=2)
assert_almost_equal(0, xt.mean(), decimal=1)
assert_almost_equal(1, xt.std(), decimal=1)
def test_empty(self):
assert_(stats.yeojohnson([]).shape == (0,))
def test_array_like(self):
np.random.seed(54321)
x = stats.norm.rvs(size=100, loc=0)
lmbda = 1.5
xt1, _ = stats.yeojohnson(x)
xt2, _ = stats.yeojohnson(list(x))
assert_allclose(xt1, xt2, rtol=1e-12)
class TestYeojohnsonNormmax(object):
def setup_method(self):
np.random.seed(12345)
self.x = stats.loggamma.rvs(5, size=50) + 5
def test_mle(self):
maxlog = stats.yeojohnson_normmax(self.x)
assert_allclose(maxlog, 1.876393, rtol=1e-6)
def test_darwin_example(self):
# test from original paper "A new family of power transformations to
# improve normality or symmetry" by Yeo and Johnson.
x = [6.1, -8.4, 1.0, 2.0, 0.7, 2.9, 3.5, 5.1, 1.8, 3.6, 7.0, 3.0, 9.3,
7.5, -6.0]
lmbda = stats.yeojohnson_normmax(x)
assert np.allclose(lmbda, 1.305, atol=1e-3)
class TestCircFuncs(object):
def test_circfuncs(self):
x = np.array([355, 5, 2, 359, 10, 350])
M = stats.circmean(x, high=360)
Mval = 0.167690146
assert_allclose(M, Mval, rtol=1e-7)
V = stats.circvar(x, high=360)
Vval = 42.51955609
assert_allclose(V, Vval, rtol=1e-7)
S = stats.circstd(x, high=360)
Sval = 6.520702116
assert_allclose(S, Sval, rtol=1e-7)
def test_circfuncs_small(self):
x = np.array([20, 21, 22, 18, 19, 20.5, 19.2])
M1 = x.mean()
M2 = stats.circmean(x, high=360)
assert_allclose(M2, M1, rtol=1e-5)
V1 = x.var()
V2 = stats.circvar(x, high=360)
assert_allclose(V2, V1, rtol=1e-4)
S1 = x.std()
S2 = stats.circstd(x, high=360)
assert_allclose(S2, S1, rtol=1e-4)
def test_circmean_axis(self):
x = np.array([[355, 5, 2, 359, 10, 350],
[351, 7, 4, 352, 9, 349],
[357, 9, 8, 358, 4, 356]])
M1 = stats.circmean(x, high=360)
M2 = stats.circmean(x.ravel(), high=360)
assert_allclose(M1, M2, rtol=1e-14)
M1 = stats.circmean(x, high=360, axis=1)
M2 = [stats.circmean(x[i], high=360) for i in range(x.shape[0])]
assert_allclose(M1, M2, rtol=1e-14)
M1 = stats.circmean(x, high=360, axis=0)
M2 = [stats.circmean(x[:, i], high=360) for i in range(x.shape[1])]
assert_allclose(M1, M2, rtol=1e-14)
def test_circvar_axis(self):
x = np.array([[355, 5, 2, 359, 10, 350],
[351, 7, 4, 352, 9, 349],
[357, 9, 8, 358, 4, 356]])
V1 = stats.circvar(x, high=360)
V2 = stats.circvar(x.ravel(), high=360)
assert_allclose(V1, V2, rtol=1e-11)
V1 = stats.circvar(x, high=360, axis=1)
V2 = [stats.circvar(x[i], high=360) for i in range(x.shape[0])]
assert_allclose(V1, V2, rtol=1e-11)
V1 = stats.circvar(x, high=360, axis=0)
V2 = [stats.circvar(x[:, i], high=360) for i in range(x.shape[1])]
assert_allclose(V1, V2, rtol=1e-11)
def test_circstd_axis(self):
x = np.array([[355, 5, 2, 359, 10, 350],
[351, 7, 4, 352, 9, 349],
[357, 9, 8, 358, 4, 356]])
S1 = stats.circstd(x, high=360)
S2 = stats.circstd(x.ravel(), high=360)
assert_allclose(S1, S2, rtol=1e-11)
S1 = stats.circstd(x, high=360, axis=1)
S2 = [stats.circstd(x[i], high=360) for i in range(x.shape[0])]
assert_allclose(S1, S2, rtol=1e-11)
S1 = stats.circstd(x, high=360, axis=0)
S2 = [stats.circstd(x[:, i], high=360) for i in range(x.shape[1])]
assert_allclose(S1, S2, rtol=1e-11)
def test_circfuncs_array_like(self):
x = [355, 5, 2, 359, 10, 350]
assert_allclose(stats.circmean(x, high=360), 0.167690146, rtol=1e-7)
assert_allclose(stats.circvar(x, high=360), 42.51955609, rtol=1e-7)
assert_allclose(stats.circstd(x, high=360), 6.520702116, rtol=1e-7)
def test_empty(self):
assert_(np.isnan(stats.circmean([])))
assert_(np.isnan(stats.circstd([])))
assert_(np.isnan(stats.circvar([])))
def test_circmean_scalar(self):
x = 1.
M1 = x
M2 = stats.circmean(x)
assert_allclose(M2, M1, rtol=1e-5)
def test_circmean_range(self):
# regression test for gh-6420: circmean(..., high, low) must be
# between `high` and `low`
m = stats.circmean(np.arange(0, 2, 0.1), np.pi, -np.pi)
assert_(m < np.pi)
assert_(m > -np.pi)
def test_circfuncs_unit8(self):
# regression test for gh-7255: overflow when working with
# numpy uint8 data type
x = np.array([150, 10], dtype='uint8')
assert_equal(stats.circmean(x, high=180), 170.0)
assert_allclose(stats.circvar(x, high=180), 437.45871686, rtol=1e-7)
assert_allclose(stats.circstd(x, high=180), 20.91551378, rtol=1e-7)
class TestMedianTest(object):
def test_bad_n_samples(self):
# median_test requires at least two samples.
assert_raises(ValueError, stats.median_test, [1, 2, 3])
def test_empty_sample(self):
# Each sample must contain at least one value.
assert_raises(ValueError, stats.median_test, [], [1, 2, 3])
def test_empty_when_ties_ignored(self):
# The grand median is 1, and all values in the first argument are
# equal to the grand median. With ties="ignore", those values are
# ignored, which results in the first sample being (in effect) empty.
# This should raise a ValueError.
assert_raises(ValueError, stats.median_test,
[1, 1, 1, 1], [2, 0, 1], [2, 0], ties="ignore")
def test_empty_contingency_row(self):
# The grand median is 1, and with the default ties="below", all the
# values in the samples are counted as being below the grand median.
# This would result a row of zeros in the contingency table, which is
# an error.
assert_raises(ValueError, stats.median_test, [1, 1, 1], [1, 1, 1])
# With ties="above", all the values are counted as above the
# grand median.
assert_raises(ValueError, stats.median_test, [1, 1, 1], [1, 1, 1],
ties="above")
def test_bad_ties(self):
assert_raises(ValueError, stats.median_test, [1, 2, 3], [4, 5],
ties="foo")
def test_bad_nan_policy(self):
assert_raises(ValueError, stats.median_test, [1, 2, 3], [4, 5], nan_policy='foobar')
def test_bad_keyword(self):
assert_raises(TypeError, stats.median_test, [1, 2, 3], [4, 5],
foo="foo")
def test_simple(self):
x = [1, 2, 3]
y = [1, 2, 3]
stat, p, med, tbl = stats.median_test(x, y)
# The median is floating point, but this equality test should be safe.
assert_equal(med, 2.0)
assert_array_equal(tbl, [[1, 1], [2, 2]])
# The expected values of the contingency table equal the contingency
# table, so the statistic should be 0 and the p-value should be 1.
assert_equal(stat, 0)
assert_equal(p, 1)
def test_ties_options(self):
# Test the contingency table calculation.
x = [1, 2, 3, 4]
y = [5, 6]
z = [7, 8, 9]
# grand median is 5.
# Default 'ties' option is "below".
stat, p, m, tbl = stats.median_test(x, y, z)
assert_equal(m, 5)
assert_equal(tbl, [[0, 1, 3], [4, 1, 0]])
stat, p, m, tbl = stats.median_test(x, y, z, ties="ignore")
assert_equal(m, 5)
assert_equal(tbl, [[0, 1, 3], [4, 0, 0]])
stat, p, m, tbl = stats.median_test(x, y, z, ties="above")
assert_equal(m, 5)
assert_equal(tbl, [[0, 2, 3], [4, 0, 0]])
def test_nan_policy_options(self):
x = [1, 2, np.nan]
y = [4, 5, 6]
mt1 = stats.median_test(x, y, nan_policy='propagate')
s, p, m, t = stats.median_test(x, y, nan_policy='omit')
assert_equal(mt1, (np.nan, np.nan, np.nan, None))
assert_allclose(s, 0.31250000000000006)
assert_allclose(p, 0.57615012203057869)
assert_equal(m, 4.0)
assert_equal(t, np.array([[0, 2],[2, 1]]))
assert_raises(ValueError, stats.median_test, x, y, nan_policy='raise')
def test_basic(self):
# median_test calls chi2_contingency to compute the test statistic
# and p-value. Make sure it hasn't screwed up the call...
x = [1, 2, 3, 4, 5]
y = [2, 4, 6, 8]
stat, p, m, tbl = stats.median_test(x, y)
assert_equal(m, 4)
assert_equal(tbl, [[1, 2], [4, 2]])
exp_stat, exp_p, dof, e = stats.chi2_contingency(tbl)
assert_allclose(stat, exp_stat)
assert_allclose(p, exp_p)
stat, p, m, tbl = stats.median_test(x, y, lambda_=0)
assert_equal(m, 4)
assert_equal(tbl, [[1, 2], [4, 2]])
exp_stat, exp_p, dof, e = stats.chi2_contingency(tbl, lambda_=0)
assert_allclose(stat, exp_stat)
assert_allclose(p, exp_p)
stat, p, m, tbl = stats.median_test(x, y, correction=False)
assert_equal(m, 4)
assert_equal(tbl, [[1, 2], [4, 2]])
exp_stat, exp_p, dof, e = stats.chi2_contingency(tbl, correction=False)
assert_allclose(stat, exp_stat)
assert_allclose(p, exp_p)
| bsd-3-clause |
wangsharp/trading-with-python | spreadApp/makeDist.py | 77 | 1720 | from distutils.core import setup
import py2exe
manifest_template = '''
<?xml version="1.0" encoding="UTF-8" standalone="yes"?>
<assembly xmlns="urn:schemas-microsoft-com:asm.v1" manifestVersion="1.0">
<assemblyIdentity
version="5.0.0.0"
processorArchitecture="x86"
name="%(prog)s"
type="win32"
/>
<description>%(prog)s Program</description>
<dependency>
<dependentAssembly>
<assemblyIdentity
type="win32"
name="Microsoft.Windows.Common-Controls"
version="6.0.0.0"
processorArchitecture="X86"
publicKeyToken="6595b64144ccf1df"
language="*"
/>
</dependentAssembly>
</dependency>
</assembly>
'''
RT_MANIFEST = 24
import matplotlib
opts = {
'py2exe': {
"compressed": 1,
"bundle_files" : 3,
"includes" : ["sip",
"matplotlib.backends",
"matplotlib.backends.backend_qt4agg",
"pylab", "numpy",
"matplotlib.backends.backend_tkagg"],
'excludes': ['_gtkagg', '_tkagg', '_agg2',
'_cairo', '_cocoaagg',
'_fltkagg', '_gtk', '_gtkcairo', ],
'dll_excludes': ['libgdk-win32-2.0-0.dll',
'libgobject-2.0-0.dll']
}
}
setup(name="triton",
version = "0.1",
scripts=["spreadScanner.pyw"],
windows=[{"script": "spreadScanner.pyw"}],
options=opts,
data_files=matplotlib.get_py2exe_datafiles(),
other_resources = [(RT_MANIFEST, 1, manifest_template % dict(prog="spreadDetective"))],
zipfile = None) | bsd-3-clause |
gfyoung/pandas | pandas/tests/groupby/test_bin_groupby.py | 2 | 3023 | import numpy as np
import pytest
from pandas._libs import lib, reduction as libreduction
import pandas as pd
from pandas import Series
import pandas._testing as tm
def test_series_grouper():
obj = Series(np.random.randn(10))
dummy = obj.iloc[:0]
labels = np.array([-1, -1, -1, 0, 0, 0, 1, 1, 1, 1], dtype=np.int64)
grouper = libreduction.SeriesGrouper(obj, np.mean, labels, 2, dummy)
result, counts = grouper.get_result()
expected = np.array([obj[3:6].mean(), obj[6:].mean()])
tm.assert_almost_equal(result, expected)
exp_counts = np.array([3, 4], dtype=np.int64)
tm.assert_almost_equal(counts, exp_counts)
def test_series_grouper_requires_nonempty_raises():
# GH#29500
obj = Series(np.random.randn(10))
dummy = obj.iloc[:0]
labels = np.array([-1, -1, -1, 0, 0, 0, 1, 1, 1, 1], dtype=np.int64)
with pytest.raises(ValueError, match="SeriesGrouper requires non-empty `series`"):
libreduction.SeriesGrouper(dummy, np.mean, labels, 2, dummy)
def test_series_bin_grouper():
obj = Series(np.random.randn(10))
dummy = obj[:0]
bins = np.array([3, 6])
grouper = libreduction.SeriesBinGrouper(obj, np.mean, bins, dummy)
result, counts = grouper.get_result()
expected = np.array([obj[:3].mean(), obj[3:6].mean(), obj[6:].mean()])
tm.assert_almost_equal(result, expected)
exp_counts = np.array([3, 3, 4], dtype=np.int64)
tm.assert_almost_equal(counts, exp_counts)
def assert_block_lengths(x):
assert len(x) == len(x._mgr.blocks[0].mgr_locs)
return 0
def cumsum_max(x):
x.cumsum().max()
return 0
@pytest.mark.parametrize("func", [cumsum_max, assert_block_lengths])
def test_mgr_locs_updated(func):
# https://github.com/pandas-dev/pandas/issues/31802
# Some operations may require creating new blocks, which requires
# valid mgr_locs
df = pd.DataFrame({"A": ["a", "a", "a"], "B": ["a", "b", "b"], "C": [1, 1, 1]})
result = df.groupby(["A", "B"]).agg(func)
expected = pd.DataFrame(
{"C": [0, 0]},
index=pd.MultiIndex.from_product([["a"], ["a", "b"]], names=["A", "B"]),
)
tm.assert_frame_equal(result, expected)
@pytest.mark.parametrize(
"binner,closed,expected",
[
(
np.array([0, 3, 6, 9], dtype=np.int64),
"left",
np.array([2, 5, 6], dtype=np.int64),
),
(
np.array([0, 3, 6, 9], dtype=np.int64),
"right",
np.array([3, 6, 6], dtype=np.int64),
),
(np.array([0, 3, 6], dtype=np.int64), "left", np.array([2, 5], dtype=np.int64)),
(
np.array([0, 3, 6], dtype=np.int64),
"right",
np.array([3, 6], dtype=np.int64),
),
],
)
def test_generate_bins(binner, closed, expected):
values = np.array([1, 2, 3, 4, 5, 6], dtype=np.int64)
result = lib.generate_bins_dt64(values, binner, closed=closed)
tm.assert_numpy_array_equal(result, expected)
class TestMoments:
pass
| bsd-3-clause |
fabioticconi/scikit-learn | examples/linear_model/plot_lasso_coordinate_descent_path.py | 42 | 2944 | """
=====================
Lasso and Elastic Net
=====================
Lasso and elastic net (L1 and L2 penalisation) implemented using a
coordinate descent.
The coefficients can be forced to be positive.
"""
print(__doc__)
# Author: Alexandre Gramfort <alexandre.gramfort@inria.fr>
# License: BSD 3 clause
from itertools import cycle
import numpy as np
import matplotlib.pyplot as plt
from sklearn.linear_model import lasso_path, enet_path
from sklearn import datasets
diabetes = datasets.load_diabetes()
X = diabetes.data
y = diabetes.target
X /= X.std(axis=0) # Standardize data (easier to set the l1_ratio parameter)
# Compute paths
eps = 5e-3 # the smaller it is the longer is the path
print("Computing regularization path using the lasso...")
alphas_lasso, coefs_lasso, _ = lasso_path(X, y, eps, fit_intercept=False)
print("Computing regularization path using the positive lasso...")
alphas_positive_lasso, coefs_positive_lasso, _ = lasso_path(
X, y, eps, positive=True, fit_intercept=False)
print("Computing regularization path using the elastic net...")
alphas_enet, coefs_enet, _ = enet_path(
X, y, eps=eps, l1_ratio=0.8, fit_intercept=False)
print("Computing regularization path using the positve elastic net...")
alphas_positive_enet, coefs_positive_enet, _ = enet_path(
X, y, eps=eps, l1_ratio=0.8, positive=True, fit_intercept=False)
# Display results
plt.figure(1)
ax = plt.gca()
colors = cycle(['b', 'r', 'g', 'c', 'k'])
neg_log_alphas_lasso = -np.log10(alphas_lasso)
neg_log_alphas_enet = -np.log10(alphas_enet)
for coef_l, coef_e, c in zip(coefs_lasso, coefs_enet, colors):
l1 = plt.plot(neg_log_alphas_lasso, coef_l, c=c)
l2 = plt.plot(neg_log_alphas_enet, coef_e, linestyle='--', c=c)
plt.xlabel('-Log(alpha)')
plt.ylabel('coefficients')
plt.title('Lasso and Elastic-Net Paths')
plt.legend((l1[-1], l2[-1]), ('Lasso', 'Elastic-Net'), loc='lower left')
plt.axis('tight')
plt.figure(2)
ax = plt.gca()
neg_log_alphas_positive_lasso = -np.log10(alphas_positive_lasso)
for coef_l, coef_pl, c in zip(coefs_lasso, coefs_positive_lasso, colors):
l1 = plt.plot(neg_log_alphas_lasso, coef_l, c=c)
l2 = plt.plot(neg_log_alphas_positive_lasso, coef_pl, linestyle='--', c=c)
plt.xlabel('-Log(alpha)')
plt.ylabel('coefficients')
plt.title('Lasso and positive Lasso')
plt.legend((l1[-1], l2[-1]), ('Lasso', 'positive Lasso'), loc='lower left')
plt.axis('tight')
plt.figure(3)
ax = plt.gca()
neg_log_alphas_positive_enet = -np.log10(alphas_positive_enet)
for (coef_e, coef_pe, c) in zip(coefs_enet, coefs_positive_enet, colors):
l1 = plt.plot(neg_log_alphas_enet, coef_e, c=c)
l2 = plt.plot(neg_log_alphas_positive_enet, coef_pe, linestyle='--', c=c)
plt.xlabel('-Log(alpha)')
plt.ylabel('coefficients')
plt.title('Elastic-Net and positive Elastic-Net')
plt.legend((l1[-1], l2[-1]), ('Elastic-Net', 'positive Elastic-Net'),
loc='lower left')
plt.axis('tight')
plt.show()
| bsd-3-clause |
Obus/scikit-learn | doc/datasets/mldata_fixture.py | 367 | 1183 | """Fixture module to skip the datasets loading when offline
Mock urllib2 access to mldata.org and create a temporary data folder.
"""
from os import makedirs
from os.path import join
import numpy as np
import tempfile
import shutil
from sklearn import datasets
from sklearn.utils.testing import install_mldata_mock
from sklearn.utils.testing import uninstall_mldata_mock
def globs(globs):
# Create a temporary folder for the data fetcher
global custom_data_home
custom_data_home = tempfile.mkdtemp()
makedirs(join(custom_data_home, 'mldata'))
globs['custom_data_home'] = custom_data_home
return globs
def setup_module():
# setup mock urllib2 module to avoid downloading from mldata.org
install_mldata_mock({
'mnist-original': {
'data': np.empty((70000, 784)),
'label': np.repeat(np.arange(10, dtype='d'), 7000),
},
'iris': {
'data': np.empty((150, 4)),
},
'datasets-uci-iris': {
'double0': np.empty((150, 4)),
'class': np.empty((150,)),
},
})
def teardown_module():
uninstall_mldata_mock()
shutil.rmtree(custom_data_home)
| bsd-3-clause |
markcheno/trading-with-python | lib/classes.py | 76 | 7847 | """
worker classes
@author: Jev Kuznetsov
Licence: GPL v2
"""
__docformat__ = 'restructuredtext'
import os
import logger as logger
import yahooFinance as yahoo
from functions import returns, rank
from datetime import date
from pandas import DataFrame, Series
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
class Symbol(object):
'''
Symbol class, the foundation of Trading With Python library,
This class acts as an interface to Yahoo data, Interactive Brokers etc
'''
def __init__(self,name):
self.name = name
self.log = logger.getLogger(self.name)
self.log.debug('class created.')
self.dataDir = os.getenv("USERPROFILE")+'\\twpData\\symbols\\'+self.name
self.log.debug('Data dir:'+self.dataDir)
self.ohlc = None # historic OHLC data
def downloadHistData(self, startDate=(2010,1,1),endDate=date.today().timetuple()[:3],\
source = 'yahoo'):
'''
get historical OHLC data from a data source (yahoo is default)
startDate and endDate are tuples in form (d,m,y)
'''
self.log.debug('Getting OHLC data')
self.ohlc = yahoo.getHistoricData(self.name,startDate,endDate)
def histData(self,column='adj_close'):
'''
Return a column of historic data.
Returns
-------------
df : DataFrame
'''
s = self.ohlc[column]
return DataFrame(s.values,s.index,[self.name])
@property
def dayReturns(self):
''' close-close returns '''
return (self.ohlc['adj_close']/self.ohlc['adj_close'].shift(1)-1)
#return DataFrame(s.values,s.index,[self.name])
class Portfolio(object):
def __init__(self,histPrice,name=''):
"""
Constructor
Parameters
----------
histPrice : historic price
"""
self.histPrice = histPrice
self.params = DataFrame(index=self.symbols)
self.params['capital'] = 100*np.ones(self.histPrice.shape[1],dtype=np.float)
self.params['last'] = self.histPrice.tail(1).T.ix[:,0]
self.params['shares'] = self.params['capital']/self.params['last']
self.name= name
def setHistPrice(self,histPrice):
self.histPrice = histPrice
def setShares(self,shares):
""" set number of shares, adjust capital
shares: list, np array or Series
"""
if len(shares) != self.histPrice.shape[1]:
raise AttributeError('Wrong size of shares vector.')
self.params['shares'] = shares
self.params['capital'] = self.params['shares']*self.params['last']
def setCapital(self,capital):
""" Set target captial, adjust number of shares """
if len(capital) != self.histPrice.shape[1]:
raise AttributeError('Wrong size of shares vector.')
self.params['capital'] = capital
self.params['shares'] = self.params['capital']/self.params['last']
def calculateStatistics(self,other=None):
''' calculate spread statistics, save internally '''
res = {}
res['micro'] = rank(self.returns[-1],self.returns)
res['macro'] = rank(self.value[-1], self.value)
res['last'] = self.value[-1]
if other is not None:
res['corr'] = self.returns.corr(returns(other))
return Series(res,name=self.name)
@property
def symbols(self):
return self.histPrice.columns.tolist()
@property
def returns(self):
return (returns(self.histPrice)*self.params['capital']).sum(axis=1)
@property
def value(self):
return (self.histPrice*self.params['shares']).sum(axis=1)
def __repr__(self):
return ("Portfolio %s \n" % self.name ) + str(self.params)
#return ('Spread %s :' % self.name ) + str.join(',',
# ['%s*%.2f' % t for t in zip(self.symbols,self.capital)])
class Spread(object):
'''
Spread class, used to build a spread out of two symbols.
'''
def __init__(self,stock,hedge,beta=None):
''' init with symbols or price series '''
if isinstance(stock,str) and isinstance(hedge,str):
self.symbols = [stock,hedge]
self._getYahooData()
elif isinstance(stock,pd.Series) and isinstance(hedge,pd.Series):
self.symbols = [stock.name,hedge.name]
self.price = pd.DataFrame(dict(zip(self.symbols,[stock,hedge]))).dropna()
else:
raise ValueError('Both stock and hedge should be of the same type, symbol string or Series')
# calculate returns
self.returns = self.price.pct_change()
if beta is not None:
self.beta = beta
else:
self.estimateBeta()
# set data
self.data = pd.DataFrame(index = self.symbols)
self.data['beta'] = pd.Series({self.symbols[0]:1., self.symbols[1]:-self.beta})
def calculateShares(self,bet):
''' set number of shares based on last quote '''
if 'price' not in self.data.columns:
print 'Getting quote...'
self.getQuote()
self.data['shares'] = bet*self.data['beta']/self.data['price']
def estimateBeta(self,plotOn=False):
""" linear estimation of beta """
x = self.returns[self.symbols[1]] # hedge
y = self.returns[self.symbols[0]] # stock
#avoid extremes
low = np.percentile(x,20)
high = np.percentile(x,80)
iValid = (x>low) & (x<high)
x = x[iValid]
y = y[iValid]
if plotOn:
plt.plot(x,y,'o')
plt.grid(True)
iteration = 1
nrOutliers = 1
while iteration < 3 and nrOutliers > 0 :
(a,b) = np.polyfit(x,y,1)
yf = np.polyval([a,b],x)
err = yf-y
idxOutlier = abs(err) > 3*np.std(err)
nrOutliers =sum(idxOutlier)
beta = a
#print 'Iteration: %i beta: %.2f outliers: %i' % (iteration,beta, nrOutliers)
x = x[~idxOutlier]
y = y[~idxOutlier]
iteration += 1
if plotOn:
yf = x*beta
plt.plot(x,yf,'-',color='red')
plt.xlabel(self.symbols[1])
plt.ylabel(self.symbols[0])
self.beta = beta
return beta
@property
def spread(self):
''' return daily returns of the pair '''
return (self.returns*self.data['beta']).sum(1)
def getQuote(self):
''' get current quote from yahoo '''
q = yahoo.getQuote(self.symbols)
self.data['price'] = q['last']
def _getYahooData(self, startDate=(2007,1,1)):
""" fetch historic data """
data = {}
for symbol in self.symbols:
print 'Downloading %s' % symbol
data[symbol]=(yahoo.getHistoricData(symbol,sDate=startDate)['adj_close'] )
self.price = pd.DataFrame(data).dropna()
def __repr__(self):
return 'Spread 1*%s & %.2f*%s ' % (self.symbols[0],-self.beta,self.symbols[1])
@property
def name(self):
return str.join('_',self.symbols)
if __name__=='__main__':
s = Spread(['SPY','IWM'])
| bsd-3-clause |
jjx02230808/project0223 | sklearn/ensemble/partial_dependence.py | 251 | 15097 | """Partial dependence plots for tree ensembles. """
# Authors: Peter Prettenhofer
# License: BSD 3 clause
from itertools import count
import numbers
import numpy as np
from scipy.stats.mstats import mquantiles
from ..utils.extmath import cartesian
from ..externals.joblib import Parallel, delayed
from ..externals import six
from ..externals.six.moves import map, range, zip
from ..utils import check_array
from ..tree._tree import DTYPE
from ._gradient_boosting import _partial_dependence_tree
from .gradient_boosting import BaseGradientBoosting
def _grid_from_X(X, percentiles=(0.05, 0.95), grid_resolution=100):
"""Generate a grid of points based on the ``percentiles of ``X``.
The grid is generated by placing ``grid_resolution`` equally
spaced points between the ``percentiles`` of each column
of ``X``.
Parameters
----------
X : ndarray
The data
percentiles : tuple of floats
The percentiles which are used to construct the extreme
values of the grid axes.
grid_resolution : int
The number of equally spaced points that are placed
on the grid.
Returns
-------
grid : ndarray
All data points on the grid; ``grid.shape[1] == X.shape[1]``
and ``grid.shape[0] == grid_resolution * X.shape[1]``.
axes : seq of ndarray
The axes with which the grid has been created.
"""
if len(percentiles) != 2:
raise ValueError('percentile must be tuple of len 2')
if not all(0. <= x <= 1. for x in percentiles):
raise ValueError('percentile values must be in [0, 1]')
axes = []
for col in range(X.shape[1]):
uniques = np.unique(X[:, col])
if uniques.shape[0] < grid_resolution:
# feature has low resolution use unique vals
axis = uniques
else:
emp_percentiles = mquantiles(X, prob=percentiles, axis=0)
# create axis based on percentiles and grid resolution
axis = np.linspace(emp_percentiles[0, col],
emp_percentiles[1, col],
num=grid_resolution, endpoint=True)
axes.append(axis)
return cartesian(axes), axes
def partial_dependence(gbrt, target_variables, grid=None, X=None,
percentiles=(0.05, 0.95), grid_resolution=100):
"""Partial dependence of ``target_variables``.
Partial dependence plots show the dependence between the joint values
of the ``target_variables`` and the function represented
by the ``gbrt``.
Read more in the :ref:`User Guide <partial_dependence>`.
Parameters
----------
gbrt : BaseGradientBoosting
A fitted gradient boosting model.
target_variables : array-like, dtype=int
The target features for which the partial dependecy should be
computed (size should be smaller than 3 for visual renderings).
grid : array-like, shape=(n_points, len(target_variables))
The grid of ``target_variables`` values for which the
partial dependecy should be evaluated (either ``grid`` or ``X``
must be specified).
X : array-like, shape=(n_samples, n_features)
The data on which ``gbrt`` was trained. It is used to generate
a ``grid`` for the ``target_variables``. The ``grid`` comprises
``grid_resolution`` equally spaced points between the two
``percentiles``.
percentiles : (low, high), default=(0.05, 0.95)
The lower and upper percentile used create the extreme values
for the ``grid``. Only if ``X`` is not None.
grid_resolution : int, default=100
The number of equally spaced points on the ``grid``.
Returns
-------
pdp : array, shape=(n_classes, n_points)
The partial dependence function evaluated on the ``grid``.
For regression and binary classification ``n_classes==1``.
axes : seq of ndarray or None
The axes with which the grid has been created or None if
the grid has been given.
Examples
--------
>>> samples = [[0, 0, 2], [1, 0, 0]]
>>> labels = [0, 1]
>>> from sklearn.ensemble import GradientBoostingClassifier
>>> gb = GradientBoostingClassifier(random_state=0).fit(samples, labels)
>>> kwargs = dict(X=samples, percentiles=(0, 1), grid_resolution=2)
>>> partial_dependence(gb, [0], **kwargs) # doctest: +SKIP
(array([[-4.52..., 4.52...]]), [array([ 0., 1.])])
"""
if not isinstance(gbrt, BaseGradientBoosting):
raise ValueError('gbrt has to be an instance of BaseGradientBoosting')
if gbrt.estimators_.shape[0] == 0:
raise ValueError('Call %s.fit before partial_dependence' %
gbrt.__class__.__name__)
if (grid is None and X is None) or (grid is not None and X is not None):
raise ValueError('Either grid or X must be specified')
target_variables = np.asarray(target_variables, dtype=np.int32,
order='C').ravel()
if any([not (0 <= fx < gbrt.n_features) for fx in target_variables]):
raise ValueError('target_variables must be in [0, %d]'
% (gbrt.n_features - 1))
if X is not None:
X = check_array(X, dtype=DTYPE, order='C')
grid, axes = _grid_from_X(X[:, target_variables], percentiles,
grid_resolution)
else:
assert grid is not None
# dont return axes if grid is given
axes = None
# grid must be 2d
if grid.ndim == 1:
grid = grid[:, np.newaxis]
if grid.ndim != 2:
raise ValueError('grid must be 2d but is %dd' % grid.ndim)
grid = np.asarray(grid, dtype=DTYPE, order='C')
assert grid.shape[1] == target_variables.shape[0]
n_trees_per_stage = gbrt.estimators_.shape[1]
n_estimators = gbrt.estimators_.shape[0]
pdp = np.zeros((n_trees_per_stage, grid.shape[0],), dtype=np.float64,
order='C')
for stage in range(n_estimators):
for k in range(n_trees_per_stage):
tree = gbrt.estimators_[stage, k].tree_
_partial_dependence_tree(tree, grid, target_variables,
gbrt.learning_rate, pdp[k])
return pdp, axes
def plot_partial_dependence(gbrt, X, features, feature_names=None,
label=None, n_cols=3, grid_resolution=100,
percentiles=(0.05, 0.95), n_jobs=1,
verbose=0, ax=None, line_kw=None,
contour_kw=None, **fig_kw):
"""Partial dependence plots for ``features``.
The ``len(features)`` plots are arranged in a grid with ``n_cols``
columns. Two-way partial dependence plots are plotted as contour
plots.
Read more in the :ref:`User Guide <partial_dependence>`.
Parameters
----------
gbrt : BaseGradientBoosting
A fitted gradient boosting model.
X : array-like, shape=(n_samples, n_features)
The data on which ``gbrt`` was trained.
features : seq of tuples or ints
If seq[i] is an int or a tuple with one int value, a one-way
PDP is created; if seq[i] is a tuple of two ints, a two-way
PDP is created.
feature_names : seq of str
Name of each feature; feature_names[i] holds
the name of the feature with index i.
label : object
The class label for which the PDPs should be computed.
Only if gbrt is a multi-class model. Must be in ``gbrt.classes_``.
n_cols : int
The number of columns in the grid plot (default: 3).
percentiles : (low, high), default=(0.05, 0.95)
The lower and upper percentile used to create the extreme values
for the PDP axes.
grid_resolution : int, default=100
The number of equally spaced points on the axes.
n_jobs : int
The number of CPUs to use to compute the PDs. -1 means 'all CPUs'.
Defaults to 1.
verbose : int
Verbose output during PD computations. Defaults to 0.
ax : Matplotlib axis object, default None
An axis object onto which the plots will be drawn.
line_kw : dict
Dict with keywords passed to the ``pylab.plot`` call.
For one-way partial dependence plots.
contour_kw : dict
Dict with keywords passed to the ``pylab.plot`` call.
For two-way partial dependence plots.
fig_kw : dict
Dict with keywords passed to the figure() call.
Note that all keywords not recognized above will be automatically
included here.
Returns
-------
fig : figure
The Matplotlib Figure object.
axs : seq of Axis objects
A seq of Axis objects, one for each subplot.
Examples
--------
>>> from sklearn.datasets import make_friedman1
>>> from sklearn.ensemble import GradientBoostingRegressor
>>> X, y = make_friedman1()
>>> clf = GradientBoostingRegressor(n_estimators=10).fit(X, y)
>>> fig, axs = plot_partial_dependence(clf, X, [0, (0, 1)]) #doctest: +SKIP
...
"""
import matplotlib.pyplot as plt
from matplotlib import transforms
from matplotlib.ticker import MaxNLocator
from matplotlib.ticker import ScalarFormatter
if not isinstance(gbrt, BaseGradientBoosting):
raise ValueError('gbrt has to be an instance of BaseGradientBoosting')
if gbrt.estimators_.shape[0] == 0:
raise ValueError('Call %s.fit before partial_dependence' %
gbrt.__class__.__name__)
# set label_idx for multi-class GBRT
if hasattr(gbrt, 'classes_') and np.size(gbrt.classes_) > 2:
if label is None:
raise ValueError('label is not given for multi-class PDP')
label_idx = np.searchsorted(gbrt.classes_, label)
if gbrt.classes_[label_idx] != label:
raise ValueError('label %s not in ``gbrt.classes_``' % str(label))
else:
# regression and binary classification
label_idx = 0
X = check_array(X, dtype=DTYPE, order='C')
if gbrt.n_features != X.shape[1]:
raise ValueError('X.shape[1] does not match gbrt.n_features')
if line_kw is None:
line_kw = {'color': 'green'}
if contour_kw is None:
contour_kw = {}
# convert feature_names to list
if feature_names is None:
# if not feature_names use fx indices as name
feature_names = [str(i) for i in range(gbrt.n_features)]
elif isinstance(feature_names, np.ndarray):
feature_names = feature_names.tolist()
def convert_feature(fx):
if isinstance(fx, six.string_types):
try:
fx = feature_names.index(fx)
except ValueError:
raise ValueError('Feature %s not in feature_names' % fx)
return fx
# convert features into a seq of int tuples
tmp_features = []
for fxs in features:
if isinstance(fxs, (numbers.Integral,) + six.string_types):
fxs = (fxs,)
try:
fxs = np.array([convert_feature(fx) for fx in fxs], dtype=np.int32)
except TypeError:
raise ValueError('features must be either int, str, or tuple '
'of int/str')
if not (1 <= np.size(fxs) <= 2):
raise ValueError('target features must be either one or two')
tmp_features.append(fxs)
features = tmp_features
names = []
try:
for fxs in features:
l = []
# explicit loop so "i" is bound for exception below
for i in fxs:
l.append(feature_names[i])
names.append(l)
except IndexError:
raise ValueError('features[i] must be in [0, n_features) '
'but was %d' % i)
# compute PD functions
pd_result = Parallel(n_jobs=n_jobs, verbose=verbose)(
delayed(partial_dependence)(gbrt, fxs, X=X,
grid_resolution=grid_resolution,
percentiles=percentiles)
for fxs in features)
# get global min and max values of PD grouped by plot type
pdp_lim = {}
for pdp, axes in pd_result:
min_pd, max_pd = pdp[label_idx].min(), pdp[label_idx].max()
n_fx = len(axes)
old_min_pd, old_max_pd = pdp_lim.get(n_fx, (min_pd, max_pd))
min_pd = min(min_pd, old_min_pd)
max_pd = max(max_pd, old_max_pd)
pdp_lim[n_fx] = (min_pd, max_pd)
# create contour levels for two-way plots
if 2 in pdp_lim:
Z_level = np.linspace(*pdp_lim[2], num=8)
if ax is None:
fig = plt.figure(**fig_kw)
else:
fig = ax.get_figure()
fig.clear()
n_cols = min(n_cols, len(features))
n_rows = int(np.ceil(len(features) / float(n_cols)))
axs = []
for i, fx, name, (pdp, axes) in zip(count(), features, names,
pd_result):
ax = fig.add_subplot(n_rows, n_cols, i + 1)
if len(axes) == 1:
ax.plot(axes[0], pdp[label_idx].ravel(), **line_kw)
else:
# make contour plot
assert len(axes) == 2
XX, YY = np.meshgrid(axes[0], axes[1])
Z = pdp[label_idx].reshape(list(map(np.size, axes))).T
CS = ax.contour(XX, YY, Z, levels=Z_level, linewidths=0.5,
colors='k')
ax.contourf(XX, YY, Z, levels=Z_level, vmax=Z_level[-1],
vmin=Z_level[0], alpha=0.75, **contour_kw)
ax.clabel(CS, fmt='%2.2f', colors='k', fontsize=10, inline=True)
# plot data deciles + axes labels
deciles = mquantiles(X[:, fx[0]], prob=np.arange(0.1, 1.0, 0.1))
trans = transforms.blended_transform_factory(ax.transData,
ax.transAxes)
ylim = ax.get_ylim()
ax.vlines(deciles, [0], 0.05, transform=trans, color='k')
ax.set_xlabel(name[0])
ax.set_ylim(ylim)
# prevent x-axis ticks from overlapping
ax.xaxis.set_major_locator(MaxNLocator(nbins=6, prune='lower'))
tick_formatter = ScalarFormatter()
tick_formatter.set_powerlimits((-3, 4))
ax.xaxis.set_major_formatter(tick_formatter)
if len(axes) > 1:
# two-way PDP - y-axis deciles + labels
deciles = mquantiles(X[:, fx[1]], prob=np.arange(0.1, 1.0, 0.1))
trans = transforms.blended_transform_factory(ax.transAxes,
ax.transData)
xlim = ax.get_xlim()
ax.hlines(deciles, [0], 0.05, transform=trans, color='k')
ax.set_ylabel(name[1])
# hline erases xlim
ax.set_xlim(xlim)
else:
ax.set_ylabel('Partial dependence')
if len(axes) == 1:
ax.set_ylim(pdp_lim[1])
axs.append(ax)
fig.subplots_adjust(bottom=0.15, top=0.7, left=0.1, right=0.95, wspace=0.4,
hspace=0.3)
return fig, axs
| bsd-3-clause |
vsoch/expfactory-docker | expdj/settings.py | 2 | 5848 | """
Django settings for expdj project.
"""
# Build paths inside the project like this: os.path.join(BASE_DIR, ...)
import os
import sys
import tempfile
from datetime import timedelta
import matplotlib
from celery import Celery
from kombu import Exchange, Queue
matplotlib.use('Agg')
BASE_DIR = os.path.dirname(os.path.dirname(__file__))
DOMAIN_NAME = "https://expfactory.org" # MUST BE HTTPS FOR MECHANICAL TURK
DOMAIN_NAME_HTTP = "http://expfactory.org" # MUST BE HTTPS FOR MECHANICAL TURK
ADMINS = (('rblair', 'rosswilsonblair@gmail.com'),)
MANAGERS = ADMINS
DEBUG = True
MTURK_ALLOW = False # Allow users to deploy to real Mturk (not just sandbox)
TEMPLATE_DEBUG = False
ALLOWED_HOSTS = ["*"]
DATABASES = {
'default': {
'ENGINE': 'django.db.backends.postgresql_psycopg2',
'NAME': 'postgres',
'USER': 'postgres',
'HOST': 'db',
'PORT': '5432',
}
}
# Application definition
INSTALLED_APPS = (
'django.contrib.admin',
'django.contrib.auth',
'django.contrib.contenttypes',
'django.contrib.sessions',
'django.contrib.messages',
'django.contrib.sites',
'django.contrib.sitemaps',
'django_user_agents',
'django.contrib.staticfiles',
'django_extensions',
'expdj.apps.main',
'expdj.apps.turk',
'expdj.apps.experiments',
'expdj.apps.users',
'crispy_forms',
'polymorphic',
'guardian',
'dbbackup',
'djcelery',
'rest_framework',
'rest_framework.authtoken',
)
AUTHENTICATION_BACKENDS = (
'django.contrib.auth.backends.ModelBackend',
'social.backends.facebook.FacebookOAuth2',
'social.backends.google.GoogleOAuth2',
'guardian.backends.ObjectPermissionBackend'
)
SOCIAL_AUTH_PIPELINE = (
'social.pipeline.social_auth.social_details',
'social.pipeline.social_auth.social_uid',
'social.pipeline.social_auth.auth_allowed',
'social.pipeline.social_auth.social_user',
'social.pipeline.user.get_username',
'social.pipeline.social_auth.associate_by_email', # <--- enable this one
'social.pipeline.user.create_user',
'social.pipeline.social_auth.associate_user',
'social.pipeline.social_auth.load_extra_data',
'social.pipeline.user.user_details'
)
SOCIAL_AUTH_FACEBOOK_SCOPE = ['email']
MIDDLEWARE_CLASSES = (
'django.contrib.sessions.middleware.SessionMiddleware',
'django.middleware.common.CommonMiddleware',
'django.middleware.csrf.CsrfViewMiddleware',
'django.contrib.auth.middleware.AuthenticationMiddleware',
'django.contrib.auth.middleware.SessionAuthenticationMiddleware',
'django.contrib.messages.middleware.MessageMiddleware',
'django.middleware.clickjacking.XFrameOptionsMiddleware',
'django_user_agents.middleware.UserAgentMiddleware',
)
ROOT_URLCONF = 'expdj.urls'
TEMPLATES = [
{
'BACKEND': 'django.template.backends.django.DjangoTemplates',
'DIRS': [os.path.join(BASE_DIR, 'templates')],
'APP_DIRS': True,
'OPTIONS': {
'context_processors': [
'django.template.context_processors.debug',
'django.template.context_processors.request',
'django.contrib.auth.context_processors.auth',
'django.contrib.messages.context_processors.messages',
],
},
},
]
TEST_RUNNER = 'django.test.runner.DiscoverRunner'
WSGI_APPLICATION = 'expdj.wsgi.application'
# Internationalization
LANGUAGE_CODE = 'en-us'
TIME_ZONE = 'UTC'
USE_I18N = True
USE_L10N = True
USE_TZ = True
SITE_ID = 1
ANONYMOUS_USER_ID = -1 # django-guardian
# Static files (CSS, JavaScript, Images)
MEDIA_ROOT = './static'
MEDIA_URL = '/static/'
STATIC_ROOT = './assets'
STATIC_URL = '/assets/'
# List of finder classes that know how to find static files in
# various locations.
STATICFILES_FINDERS = (
'django.contrib.staticfiles.finders.FileSystemFinder',
'django.contrib.staticfiles.finders.AppDirectoriesFinder',
)
SESSION_SERIALIZER = 'django.contrib.sessions.serializers.PickleSerializer'
SENDFILE_BACKEND = 'sendfile.backends.development'
PRIVATE_MEDIA_REDIRECT_HEADER = 'X-Accel-Redirect'
CRISPY_TEMPLATE_PACK = 'bootstrap3'
CACHES = {
'default': {
'BACKEND': 'django.core.cache.backends.locmem.LocMemCache',
}
}
# Celery config
BROKER_URL = 'redis://redis:6379/0'
CELERY_RESULT_BACKEND = 'djcelery.backends.database:DatabaseBackend'
CELERY_ACCEPT_CONTENT = ['json']
CELERY_TASK_SERIALIZER = 'json'
CELERY_RESULT_SERIALIZER = 'json'
CELERY_DEFAULT_QUEUE = 'default'
CELERY_QUEUES = (
Queue('default', Exchange('default'), routing_key='default'),
)
CELERY_IMPORTS = ('expdj.apps.turk.tasks', )
# here is how to run a task regularly
# CELERYBEAT_SCHEDULE = {
# 'task name': {
# 'task': 'task_name',
# 'schedule': timedelta(days=1)
# },
# }
CELERY_TIMEZONE = 'Europe/Berlin'
# REST FRAMEWORK
REST_FRAMEWORK = {
# Use Django's standard `django.contrib.auth` permissions,
# or allow read-only access for unauthenticated users.
'DEFAULT_PERMISSION_CLASSES': [
'rest_framework.permissions.IsAuthenticatedOrReadOnly',
],
'DEFAULT_AUTHENTICATION_CLASSES': (
'rest_framework.authentication.BasicAuthentication',
'rest_framework.authentication.SessionAuthentication',
'rest_framework.authentication.TokenAuthentication',
),
'DEFAULT_PAGINATION_CLASS': 'rest_framework.pagination.LimitOffsetPagination',
'PAGE_SIZE': 10,
}
CSRF_COOKIE_SECURE = False
SESSION_COOKIE_SECURE = False
# SSL ENABLED
SECURE_PROXY_SSL_HEADER = ('HTTP_X_FORWARDED_PROTOCOL', 'https')
EXP_REPO = os.path.join(BASE_DIR, 'expdj/experiment_repo')
# Bogus secret key.
try:
from secrets import *
except ImportError:
from bogus_secrets import *
# Local settings
try:
from local_settings import *
except ImportError:
pass
| mit |
datacommonsorg/data | scripts/us_bjs/nps/preprocess_data.py | 1 | 19122 | import pandas as pd
from absl import flags
from absl import app
FLAGS = flags.FLAGS
flags.DEFINE_string('preprocess_file',
'NPS_1978-2018_Data.tsv',
'file path to tsv file with data to proess',
short_name='p')
def convert_nan_for_calculation(value):
if pd.isna(value):
return 0
else:
return value
def total_jurisdiction_columns_helper(df):
"""calculation to include private facility numbers"""
df["PVINF_Temp"] = df["PVINF"].apply(convert_nan_for_calculation)
df["PVOTHF_Temp"] = df["PVOTHF"].apply(convert_nan_for_calculation)
df["PVINM_Temp"] = df["PVINM"].apply(convert_nan_for_calculation)
df["PVOTHM_Temp"] = df["PVOTHM"].apply(convert_nan_for_calculation)
df["Female_Total_Temp"] = df[["JURTOTF", "PVINF_Temp", "PVOTHF_Temp"
]].sum(axis=1).where(df["PVINCLF"] == 2,
df["JURTOTF"])
df["Male_Total_Temp"] = df[["JURTOTM", "PVINM_Temp", "PVOTHM_Temp"
]].sum(axis=1).where(df["PVINCLM"] == 2,
df["JURTOTM"])
"""calculation to include local facility numbers"""
df["LFF_Temp"] = df["LFF"].apply(convert_nan_for_calculation)
df["LFM_Temp"] = df["LFM"].apply(convert_nan_for_calculation)
df["Female_Total_Temp"] = df[["Female_Total_Temp", "LFF_Temp"
]].sum(axis=1).where(df["LFINCLF"] == 2,
df["Female_Total_Temp"])
df["Male_Total_Temp"] = df[["Male_Total_Temp", "LFM_Temp"
]].sum(axis=1).where(df["LFINCLM"] == 2,
df["Male_Total_Temp"])
"""calculation to include numbers from local facilities solely to ease crowding"""
df["LFCRSTF_Temp"] = df["LFCRSTF"].apply(convert_nan_for_calculation)
df["LFCRSTM_Temp"] = df["LFCRSTM"].apply(convert_nan_for_calculation)
df["Female_Total_Temp"] = df[["Female_Total_Temp", "LFCRSTF_Temp"
]].sum(axis=1).where(df["LFCRINCF"] == 2,
df["Female_Total_Temp"])
df["Male_Total_Temp"] = df[["Male_Total_Temp", "LFCRSTM_Temp"
]].sum(axis=1).where(df["LFCRINCM"] == 2,
df["Male_Total_Temp"])
"""calculation to include federal and other state facility numbers"""
df["FEDF_Temp"] = df["FEDF"].apply(convert_nan_for_calculation)
df["OTHSTF_Temp"] = df["OTHSTF"].apply(convert_nan_for_calculation)
df["FEDM_Temp"] = df["FEDM"].apply(convert_nan_for_calculation)
df["OTHSTM_Temp"] = df["OTHSTM"].apply(convert_nan_for_calculation)
df["Female_Total_Temp"] = df[[
"Female_Total_Temp", "FEDF_Temp", "OTHSTF_Temp"
]].sum(axis=1).where(df["FACINCLF"] == 2, df["Female_Total_Temp"])
df["Male_Total_Temp"] = df[["Male_Total_Temp", "FEDM_Temp", "OTHSTM_Temp"
]].sum(axis=1).where(df["FACINCLM"] == 2,
df["Male_Total_Temp"])
def get_columns(df):
df_out = {}
total_jurisdiction_columns_helper(df)
df_out["GeoId"] = df["GeoId"]
df_out["YEAR"] = df["YEAR"]
df_out["Count_Person_Female_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"Female_Total_Temp"]
df_out[
"Count_Person_Female_Incarcerated_WhiteAlone_MeasuredBasedOnJurisdiction"] = df[
"WHITEF"]
df_out[
"Count_Person_BlackOrAfricanAmericanAlone_Female_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"BLACKF"]
df_out[
"Count_Person_Female_HispanicOrLatino_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"HISPF"]
df_out[
"Count_Person_AmericanIndianOrAlaskaNativeAlone_Female_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"AIANF"]
df_out[
"Count_Person_AsianAlone_Female_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"ASIANF"]
df_out[
"Count_Person_Female_Incarcerated_NativeHawaiianOrOtherPacificIslanderAlone_MeasuredBasedOnJurisdiction"] = df[
"NHPIF"]
df_out[
"Count_Person_Female_Incarcerated_TwoOrMoreRaces_MeasuredBasedOnJurisdiction"] = df[
"TWORACEF"]
df_out[
"Count_MortalityEvent_Female_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHTOTF"]
df_out[
"Count_MortalityEvent_Female_Incarcerated_JudicialExecution_MeasuredBasedOnJurisdiction"] = df[
"DTHEXECF"]
df_out[
"Count_MortalityEvent_Female_IllnessOrNaturalCause_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHILLNF"]
df_out[
"Count_MortalityEvent_AIDS_Female_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHAIDSF"]
df_out[
"Count_MortalityEvent_Female_Incarcerated_IntentionalSelf-Harm(Suicide)_MeasuredBasedOnJurisdiction"] = df[
"DTHSUICF"]
df_out[
"Count_MortalityEvent_Accidents(UnintentionalInjuries)_Female_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHACCF"]
df_out[
"Count_MortalityEvent_DeathDueToAnotherPerson_Female_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHPERSF"]
df_out[
"Count_MortalityEvent_Assault(Homicide)_Female_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHHOMIF"]
df_out[
"Count_MortalityEvent_Female_Incarcerated_NPSOtherCauseOfDeath_MeasuredBasedOnJurisdiction"] = df[
"DTHOTHF"]
df_out[
"Count_IncarcerationEvent_AdmittedToPrison_Female_Incarcerated_MaxSentenceGreaterThan1Year_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"ADTOTF"]
df_out[
"Count_IncarcerationEvent_Female_Incarcerated_MaxSentenceGreaterThan1Year_ReleasedFromPrison_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"RLTOTF"]
df_out[
"Count_Person_Female_Incarcerated_MaxSentenceGreaterThan1Year_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"JURGT1F"]
df_out[
"Count_Person_Female_Incarcerated_MaxSentence1YearOrLess_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"JURLT1F"]
df_out[
"Count_Person_Female_Incarcerated_Unsentenced_MeasuredBasedOnJurisdiction"] = df[
"JURUNSF"]
df_out[
"Count_Person_Female_Incarcerated_InState_PrivatelyOperated_MeasuredBasedOnJurisdiction"] = df[
"PVINF"]
df_out[
"Count_Person_Female_Incarcerated_OutOfState_PrivatelyOperated_MeasuredBasedOnJurisdiction"] = df[
"PVOTHF"]
df_out[
"Count_Person_Female_Incarcerated_Local_LocallyOperated_MeasuredBasedOnJurisdiction"] = df[
"LFF"]
df_out[
"Count_Person_FederallyOperated_Female_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"FEDF"]
df_out[
"Count_Person_Female_Incarcerated_OutOfState_StateOperated_MeasuredBasedOnJurisdiction"] = df[
"OTHSTF"]
df_out[
"Count_Person_Female_Incarcerated_NotAUSCitizen_StateOperated&FederallyOperated&PrivatelyOperated_MeasuredBasedOnCustody"] = df[
"NCITZTOTF"]
df_out[
"Count_Person_Female_Incarcerated_MaxSentenceGreaterThan1Year_NotAUSCitizen_Sentenced_StateOperated&FederallyOperated&PrivatelyOperated_MeasuredBasedOnCustody"] = df[
"NCITZGT1F"]
df_out[
"Count_Person_Female_Incarcerated_MaxSentence1YearOrLess_NotAUSCitizen_Sentenced_StateOperated&FederallyOperated&PrivatelyOperated_MeasuredBasedOnCustody"] = df[
"NCITZLE1F"]
df_out[
"Count_Person_Female_Incarcerated_NotAUSCitizen_StateOperated&FederallyOperated&PrivatelyOperated_Unsentenced_MeasuredBasedOnCustody"] = df[
"NCITZUNSF"]
df_out[
"Count_Person_Female_Incarcerated_Under18_MeasuredBasedOnCustody"] = df[
"CUSLT18F"]
df_out["Count_Person_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"Male_Total_Temp"]
df_out[
"Count_Person_Incarcerated_Male_WhiteAlone_MeasuredBasedOnJurisdiction"] = df[
"WHITEM"]
df_out[
"Count_Person_BlackOrAfricanAmericanAlone_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"BLACKM"]
df_out[
"Count_Person_HispanicOrLatino_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"HISPM"]
df_out[
"Count_Person_AmericanIndianOrAlaskaNativeAlone_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"AIANM"]
df_out[
"Count_Person_AsianAlone_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"ASIANM"]
df_out[
"Count_Person_Incarcerated_Male_NativeHawaiianOrOtherPacificIslanderAlone_MeasuredBasedOnJurisdiction"] = df[
"NHPIM"]
df_out[
"Count_Person_Incarcerated_Male_TwoOrMoreRaces_MeasuredBasedOnJurisdiction"] = df[
"TWORACEM"]
df_out[
"Count_MortalityEvent_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"DTHTOTM"]
df_out[
"Count_MortalityEvent_Incarcerated_JudicialExecution_Male_MeasuredBasedOnJurisdiction"] = df[
"DTHEXECM"]
df_out[
"Count_MortalityEvent_IllnessOrNaturalCause_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"DTHILLNM"]
df_out[
"Count_MortalityEvent_AIDS_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"DTHAIDSM"]
df_out[
"Count_MortalityEvent_Incarcerated_IntentionalSelf-Harm(Suicide)_Male_MeasuredBasedOnJurisdiction"] = df[
"DTHSUICM"]
df_out[
"Count_MortalityEvent_Accidents(UnintentionalInjuries)_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"DTHACCM"]
df_out[
"Count_MortalityEvent_DeathDueToAnotherPerson_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"DTHPERSM"]
df_out[
"Count_MortalityEvent_Assault(Homicide)_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"DTHHOMIM"]
df_out[
"Count_MortalityEvent_Incarcerated_Male_NPSOtherCauseOfDeath_MeasuredBasedOnJurisdiction"] = df[
"DTHOTHM"]
df_out[
"Count_IncarcerationEvent_AdmittedToPrison_Incarcerated_Male_MaxSentenceGreaterThan1Year_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"ADTOTM"]
df_out[
"Count_IncarcerationEvent_Incarcerated_Male_MaxSentenceGreaterThan1Year_ReleasedFromPrison_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"RLTOTM"]
df_out[
"Count_Person_Incarcerated_Male_MaxSentenceGreaterThan1Year_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"JURGT1M"]
df_out[
"Count_Person_Incarcerated_Male_MaxSentence1YearOrLess_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"JURLT1M"]
df_out[
"Count_Person_Incarcerated_Male_Unsentenced_MeasuredBasedOnJurisdiction"] = df[
"JURUNSM"]
df_out[
"Count_Person_Incarcerated_InState_Male_PrivatelyOperated_MeasuredBasedOnJurisdiction"] = df[
"PVINM"]
df_out[
"Count_Person_Incarcerated_Male_OutOfState_PrivatelyOperated_MeasuredBasedOnJurisdiction"] = df[
"PVOTHM"]
df_out[
"Count_Person_Incarcerated_Local_LocallyOperated_Male_MeasuredBasedOnJurisdiction"] = df[
"LFM"]
df_out[
"Count_Person_FederallyOperated_Incarcerated_Male_MeasuredBasedOnJurisdiction"] = df[
"FEDM"]
df_out[
"Count_Person_Incarcerated_Male_OutOfState_StateOperated_MeasuredBasedOnJurisdiction"] = df[
"OTHSTM"]
df_out[
"Count_Person_Incarcerated_Male_NotAUSCitizen_StateOperated&FederallyOperated&PrivatelyOperated_MeasuredBasedOnCustody"] = df[
"NCITZTOTM"]
df_out[
"Count_Person_Incarcerated_Male_MaxSentenceGreaterThan1Year_NotAUSCitizen_Sentenced_StateOperated&FederallyOperated&PrivatelyOperated_MeasuredBasedOnCustody"] = df[
"NCITZGT1M"]
df_out[
"Count_Person_Incarcerated_Male_MaxSentence1YearOrLess_NotAUSCitizen_Sentenced_StateOperated&FederallyOperated&PrivatelyOperated_MeasuredBasedOnCustody"] = df[
"NCITZLE1M"]
df_out[
"Count_Person_Incarcerated_Male_NotAUSCitizen_StateOperated&FederallyOperated&PrivatelyOperated_Unsentenced_MeasuredBasedOnCustody"] = df[
"NCITZUNSM"]
df_out[
"Count_Person_Incarcerated_Male_Under18_MeasuredBasedOnCustody"] = df[
"CUSLT18M"]
df_out["Count_Person_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"Female_Total_Temp"] + df["Male_Total_Temp"]
df_out[
"Count_Person_Incarcerated_WhiteAlone_MeasuredBasedOnJurisdiction"] = df[
"WHITEF"] + df["WHITEM"]
df_out[
"Count_Person_BlackOrAfricanAmericanAlone_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"BLACKF"] + df["BLACKM"]
df_out[
"Count_Person_HispanicOrLatino_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"HISPF"] + df["HISPM"]
df_out[
"Count_Person_AmericanIndianOrAlaskaNativeAlone_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"AIANF"] + df["AIANM"]
df_out[
"Count_Person_AsianAlone_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"ASIANF"] + df["ASIANM"]
df_out[
"Count_Person_Incarcerated_NativeHawaiianOrOtherPacificIslanderAlone_MeasuredBasedOnJurisdiction"] = df[
"NHPIF"] + df["NHPIM"]
df_out[
"Count_Person_Incarcerated_TwoOrMoreRaces_MeasuredBasedOnJurisdiction"] = df[
"TWORACEF"] + df["TWORACEM"]
df_out[
"Count_MortalityEvent_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHTOTF"] + df["DTHTOTM"]
df_out[
"Count_MortalityEvent_Incarcerated_JudicialExecution_MeasuredBasedOnJurisdiction"] = df[
"DTHEXECF"] + df["DTHEXECM"]
df_out[
"Count_MortalityEvent_IllnessOrNaturalCause_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHILLNF"] + df["DTHILLNM"]
df_out[
"Count_MortalityEvent_AIDS_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHAIDSF"] + df["DTHAIDSM"]
df_out[
"Count_MortalityEvent_Incarcerated_IntentionalSelf-Harm(Suicide)_MeasuredBasedOnJurisdiction"] = df[
"DTHSUICF"] + df["DTHSUICM"]
df_out[
"Count_MortalityEvent_Accidents(UnintentionalInjuries)_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHACCF"] + df["DTHACCM"]
df_out[
"Count_MortalityEvent_DeathDueToAnotherPerson_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHPERSF"] + df["DTHPERSM"]
df_out[
"Count_MortalityEvent_Assault(Homicide)_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"DTHHOMIF"] + df["DTHHOMIM"]
df_out[
"Count_MortalityEvent_Incarcerated_NPSOtherCauseOfDeath_MeasuredBasedOnJurisdiction"] = df[
"DTHOTHF"] + df["DTHOTHM"]
df_out[
"Count_IncarcerationEvent_AdmittedToPrison_Incarcerated_MaxSentenceGreaterThan1Year_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"ADTOTF"] + df["ADTOTM"]
df_out[
"Count_IncarcerationEvent_Incarcerated_MaxSentenceGreaterThan1Year_ReleasedFromPrison_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"RLTOTF"] + df["RLTOTM"]
df_out[
"Count_Person_Incarcerated_MaxSentenceGreaterThan1Year_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"JURGT1F"] + df["JURGT1M"]
df_out[
"Count_Person_Incarcerated_MaxSentence1YearOrLess_Sentenced_MeasuredBasedOnJurisdiction"] = df[
"JURLT1F"] + df["JURLT1M"]
df_out[
"Count_Person_Incarcerated_Unsentenced_MeasuredBasedOnJurisdiction"] = df[
"JURUNSF"] + df["JURUNSM"]
df_out[
"Count_Person_Incarcerated_InState_PrivatelyOperated_MeasuredBasedOnJurisdiction"] = df[
"PVINF"] + df["PVINM"]
df_out[
"Count_Person_Incarcerated_OutOfState_PrivatelyOperated_MeasuredBasedOnJurisdiction"] = df[
"PVOTHF"] + df["PVOTHM"]
df_out[
"Count_Person_Incarcerated_Local_LocallyOperated_MeasuredBasedOnJurisdiction"] = df[
"LFF"] + df["LFM"]
df_out[
"Count_Person_FederallyOperated_Incarcerated_MeasuredBasedOnJurisdiction"] = df[
"FEDF"] + df["FEDM"]
df_out[
"Count_Person_Incarcerated_OutOfState_StateOperated_MeasuredBasedOnJurisdiction"] = df[
"OTHSTF"] + df["OTHSTM"]
df_out[
"Count_Person_Incarcerated_NotAUSCitizen_StateOperated&FederallyOperated&PrivatelyOperated_MeasuredBasedOnCustody"] = df[
"NCITZTOTF"] + df["NCITZTOTM"]
df_out[
"Count_Person_Incarcerated_MaxSentenceGreaterThan1Year_NotAUSCitizen_Sentenced_StateOperated&FederallyOperated&PrivatelyOperated_MeasuredBasedOnCustody"] = df[
"NCITZGT1F"] + df["NCITZGT1M"]
df_out[
"Count_Person_Incarcerated_MaxSentence1YearOrLess_NotAUSCitizen_Sentenced_StateOperated&FederallyOperated&PrivatelyOperated_MeasuredBasedOnCustody"] = df[
"NCITZLE1F"] + df["NCITZLE1M"]
df_out[
"Count_Person_Incarcerated_NotAUSCitizen_StateOperated&FederallyOperated&PrivatelyOperated_Unsentenced_MeasuredBasedOnCustody"] = df[
"NCITZUNSF"] + df["NCITZUNSM"]
df_out["Count_Person_Incarcerated_Under18_MeasuredBasedOnCustody"] = df[
"CUSLT18F"] + df["CUSLT18M"]
return df_out
def convert_geoId(fips_code):
"""Creates geoId column"""
return 'geoId/' + str(fips_code).zfill(2)
def convert_missing_value_to_nan(value):
"""codes for missing values are always negative and actual data is always >= 0"""
if isinstance(value, int) and value < 0:
return float("nan")
else:
return value
def convert_nan_to_empty_cell(value):
if pd.isna(value):
return ''
else:
return value
def preprocess_df(raw_df):
"""cleans raw_df
Args:
raw_data: raw data frame to be used as starting point for cleaning
"""
df = raw_df.copy()
df['GeoId'] = df['STATEID'].apply(convert_geoId)
# convert missing values to NaN for aggregation
for column_name in list(df.columns):
df[column_name] = df[column_name].apply(convert_missing_value_to_nan)
#get columns matching stat var names and add aggregate columns
df_out = pd.DataFrame(get_columns(df))
#convert NaN to empty cell
for column_name in list(df_out.columns):
df_out[column_name] = df_out[column_name].apply(
convert_nan_to_empty_cell)
return df_out
def main(args):
filename = FLAGS.preprocess_file
print('Processing {0}'.format(filename))
df = pd.read_csv(filename, delimiter='\t')
processed_df = preprocess_df(df)
processed_df.to_csv(filename.replace('.tsv', '_processed.csv'), index=False)
print('Done processing {0}'.format(filename))
if __name__ == '__main__':
app.run(main)
| apache-2.0 |
apache/incubator-mxnet | python/mxnet/numpy/random.py | 4 | 40839 | # Licensed to the Apache Software Foundation (ASF) under one
# or more contributor license agreements. See the NOTICE file
# distributed with this work for additional information
# regarding copyright ownership. The ASF licenses this file
# to you under the Apache License, Version 2.0 (the
# "License"); you may not use this file except in compliance
# with the License. You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing,
# software distributed under the License is distributed on an
# "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY
# KIND, either express or implied. See the License for the
# specific language governing permissions and limitations
# under the License.
"""Namespace for ops used in imperative programming."""
from ..ndarray import numpy as _mx_nd_np
from ..random import seed
__all__ = ["randint", "uniform", "normal", "choice", "rand", "multinomial", "multivariate_normal",
"logistic", "gumbel", "f",
"laplace",
"shuffle", "randn", "gamma", "beta", "chisquare", "exponential", "lognormal",
"weibull", "pareto", "power", "rayleigh",
"seed"]
def randint(low, high=None, size=None, dtype=None, ctx=None, out=None):
r"""Return random integers from `low` (inclusive) to `high` (exclusive).
Return random integers from the "discrete uniform" distribution of
the specified dtype in the "half-open" interval [`low`, `high`). If
`high` is None (the default), then results are from [0, `low`).
Parameters
----------
low : int
Lowest (signed) integer to be drawn from the distribution (unless
``high=None``, in which case this parameter is one above the
*highest* such integer).
high : int, optional
If provided, one above the largest (signed) integer to be drawn
from the distribution (see above for behavior if ``high=None``).
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. Default is None, in which case a
single value is returned.
dtype : dtype, optional
Desired dtype of the result. All dtypes are determined by their
name, i.e., 'int64', 'int', etc, so byteorder is not available
and a specific precision may have different C types depending
on the platform. The default value is 'np.int'.
ctx : Context, optional
Device context of output. Default is current context.
out : ndarray, optional
The output ndarray (default is `None`).
Returns
-------
out : ndarray of ints
`size`-shaped array of random integers from the appropriate
distribution, or a single such random int if `size` not provided.
Examples
--------
>>> np.random.randint(2, size=10)
array([1, 0, 0, 0, 1, 1, 0, 0, 1, 0])
>>> np.random.randint(1, size=10)
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0])
Generate a 2 x 4 array of ints between 0 and 4, inclusive:
>>> np.random.randint(5, size=(2, 4))
array([[4, 0, 2, 1],
[3, 2, 2, 0]])
"""
return _mx_nd_np.random.randint(low, high, size, dtype, ctx, out)
def uniform(low=0.0, high=1.0, size=None, dtype=None, ctx=None, out=None):
r"""Draw samples from a uniform distribution.
Samples are uniformly distributed over the half-open interval
``[low, high)`` (includes low, but excludes high). In other words,
any value within the given interval is equally likely to be drawn
by `uniform`.
Parameters
----------
low : float, ndarray, optional
Lower boundary of the output interval. All values generated will be
greater than or equal to low. The default value is 0.
high : float, ndarray, optional
Upper boundary of the output interval. All values generated will be
less than high. The default value is 1.0.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a scalar tensor containing a single value is returned if
``low`` and ``high`` are both scalars. Otherwise,
``np.broadcast(low, high).size`` samples are drawn.
dtype : {'float16', 'float32', 'float64'}, optional
Data type of output samples.
When npx.is_np_default_dtype() returns False, default dtype is float32;
When npx.is_np_default_dtype() returns True, default dtype is float64.
ctx : Context, optional
Device context of output. Default is current context.
Returns
-------
out : ndarray
Drawn samples from the parameterized uniform distribution.
See Also
--------
randint : Discrete uniform distribution, yielding integers.
rand : Convenience function that accepts dimensions as input, e.g.,
``rand(2,2)`` would generate a 2-by-2 array of floats,
uniformly distributed over ``[0, 1)``.
Notes
-----
The probability density function of the uniform distribution is
.. math:: p(x) = \frac{1}{b - a}
anywhere within the interval ``[a, b)``, and zero elsewhere.
When ``high`` == ``low``, values of ``low`` will be returned.
If ``high`` < ``low``, the results are officially undefined
and may eventually raise an error, i.e. do not rely on this
function to behave when passed arguments satisfying that
inequality condition.
"""
return _mx_nd_np.random.uniform(low, high, size=size, ctx=ctx, dtype=dtype, out=out)
def normal(loc=0.0, scale=1.0, size=None, dtype=None, ctx=None, out=None):
r"""Draw random samples from a normal (Gaussian) distribution.
Samples are distributed according to a normal distribution parametrized
by *loc* (mean) and *scale* (standard deviation).
Parameters
----------
loc : float, optional
Mean (centre) of the distribution.
scale : float, optional
Standard deviation (spread or "width") of the distribution.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., `(m, n, k)`, then `m * n * k`
samples are drawn. If size is `None` (default), a scalar tensor containing
a single value is returned if loc and scale are both scalars. Otherwise,
``np.broadcast(low, high).size`` samples are drawn.
dtype : {'float16', 'float32', 'float64'}, optional
Data type of output samples.
When npx.is_np_default_dtype() returns False, default dtype is float32;
When npx.is_np_default_dtype() returns True, default dtype is float64.
ctx : Context, optional
Device context of output, default is current context.
out : ``ndarray``, optional
Store output to an existing ``ndarray``.
Returns
-------
out : ndarray
Drawn samples from the parameterized `normal distribution` [1]_.
Notes
-----
The probability density for the Gaussian distribution is
.. math:: p(x) = \frac{1}{\sqrt{ 2 \pi \sigma^2 }}
e^{ - \frac{ (x - \mu)^2 } {2 \sigma^2} },
where :math:`\mu` is the mean and :math:`\sigma` the standard
deviation. The square of the standard deviation, :math:`\sigma^2`,
is called the variance.
The function has its peak at the mean, and its "spread" increases with
the standard deviation (the function reaches 0.607 times its maximum at
:math:`x + \sigma` and :math:`x - \sigma` [2]_). This implies that
`numpy.random.normal` is more likely to return samples lying close to
the mean, rather than those far away.
References
----------
.. [1] Wikipedia, "Normal distribution",
https://en.wikipedia.org/wiki/Normal_distribution
.. [2] P. R. Peebles Jr., "Central Limit Theorem" in "Probability,
Random Variables and Random Signal Principles", 4th ed., 2001,
pp. 51, 51, 125.
Examples
--------
>>> mu, sigma = 0, 0.1 # mean and standard deviation
>>> s = np.random.normal(mu, sigma, 1000)
Verify the mean and the variance:
>>> np.abs(mu - np.mean(s)) < 0.01
array(True)
"""
return _mx_nd_np.random.normal(loc, scale, size, dtype, ctx, out)
def lognormal(mean=0.0, sigma=1.0, size=None, dtype=None, ctx=None, out=None):
r"""Draw samples from a log-normal distribution.
Draw samples from a `log-normal distribution` [1]_ with specified mean,
standard deviation, and array shape. Note that the mean and standard
deviation are not the values for the distribution itself, but of the
underlying normal distribution it is derived from.
Parameters
----------
mean : float or array_like of floats, optional
Mean value of the underlying normal distribution. Default is 0.
sigma : float or array_like of floats, optional
Standard deviation of the underlying normal distribution. Must be
non-negative. Default is 1.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``mean`` and ``sigma`` are both scalars.
Otherwise, ``np.broadcast(mean, sigma).size`` samples are drawn.
dtype : {'float16', 'float32', 'float64'}, optional
Data type of output samples. Default is 'float32'
ctx : Context, optional
Device context of output. Default is current context.
out : ``ndarray``, optional
Store output to an existing ``ndarray``.
Returns
-------
out : ndarray or scalar
Drawn samples from the parameterized log-normal distribution.
Notes
-----
A variable `x` has a log-normal distribution if `log(x)` is normally
distributed. The `probability density function for the log-normal
distribution` [2]_ is:
.. math:: p(x) = \frac{1}{\sigma x \sqrt{2\pi}}
e^{(-\frac{(ln(x)-\mu)^2}{2\sigma^2})}
where :math:`\mu` is the mean and :math:`\sigma` is the standard
deviation of the normally distributed logarithm of the variable.
A log-normal distribution results if a random variable is the *product*
of a large number of independent, identically-distributed variables in
the same way that a normal distribution results if the variable is the
*sum* of a large number of independent, identically-distributed
variables.
References
----------
.. [1] Limpert, E., Stahel, W. A., and Abbt, M., "Log-normal
Distributions across the Sciences: Keys and Clues,"
BioScience, Vol. 51, No. 5, May, 2001.
https://stat.ethz.ch/~stahel/lognormal/bioscience.pdf
.. [2] Reiss, R.D. and Thomas, M., "Statistical Analysis of Extreme
Values," Basel: Birkhauser Verlag, 2001, pp. 31-32.
Examples
--------
Draw samples from the distribution:
>>> mu, sigma = 3., 1. # mean and standard deviation
>>> s = np.random.lognormal(mu, sigma, 1000)
"""
return _mx_nd_np.random.lognormal(mean, sigma, size, dtype, ctx, out)
def logistic(loc=0.0, scale=1.0, size=None, ctx=None, out=None):
r"""Draw samples from a logistic distribution.
Samples are drawn from a logistic distribution with specified
parameters, loc (location or mean, also median), and scale (>0).
Parameters
----------
loc : float or array_like of floats, optional
Parameter of the distribution. Default is 0.
scale : float or array_like of floats, optional
Parameter of the distribution. Must be non-negative.
Default is 1.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``loc`` and ``scale`` are both scalars.
Otherwise, ``np.broadcast(loc, scale).size`` samples are drawn.
ctx : Context, optional
Device context of output, default is current context.
out : ``ndarray``, optional
Store output to an existing ``ndarray``.
Returns
-------
out : ndarray or scalar
Drawn samples from the parameterized logistic distribution.
Examples
--------
Draw samples from the distribution:
>>> loc, scale = 10, 1
>>> s = np.random.logistic(loc, scale, 10000)
>>> import matplotlib.pyplot as plt
>>> count, bins, ignored = plt.hist(s, bins=50)
# plot against distribution
>>> def logist(x, loc, scale):
... return np.exp((loc-x)/scale)/(scale*(1+np.exp((loc-x)/scale))**2)
>>> lgst_val = logist(bins, loc, scale)
>>> plt.plot(bins, lgst_val * count.max() / lgst_val.max())
>>> plt.show()
"""
return _mx_nd_np.random.logistic(loc, scale, size, ctx, out)
def gumbel(loc=0.0, scale=1.0, size=None, ctx=None, out=None):
r"""Draw samples from a Gumbel distribution.
Draw samples from a Gumbel distribution with specified location and
scale.
Parameters
----------
loc : float or array_like of floats, optional
The location of the mode of the distribution. Default is 0.
scale : float or array_like of floats, optional
The scale parameter of the distribution. Default is 1. Must be non-
negative.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``loc`` and ``scale`` are both scalars.
Otherwise, ``np.broadcast(loc, scale).size`` samples are drawn.
ctx : Context, optional
Device context of output, default is current context.
out : ``ndarray``, optional
Store output to an existing ``ndarray``.
Returns
-------
out : ndarray or scalar
Drawn samples from the parameterized Gumbel distribution.
Examples
--------
Draw samples from the distribution:
>>> mu, beta = 0, 0.1 # location and scale
>>> s = np.random.gumbel(mu, beta, 1000)
Display the histogram of the samples, along with
the probability density function:
>>> import matplotlib.pyplot as plt
>>> count, bins, ignored = plt.hist(s, 30, density=True)
>>> plt.plot(bins, (1/beta)*np.exp(-(bins - mu)/beta)
... * np.exp( -np.exp( -(bins - mu) /beta) ),
... linewidth=2, color='r')
>>> plt.show()
Show how an extreme value distribution can arise from a Gaussian process
and compare to a Gaussian:
>>> means = []
>>> maxima = []
>>> for i in range(0,1000) :
... a = np.random.normal(mu, beta, 1000)
... means.append(a.mean())
... maxima.append(a.max())
>>> count, bins, ignored = plt.hist(maxima, 30, density=True)
>>> beta = np.std(maxima) * np.sqrt(6) / np.pi
>>> mu = np.mean(maxima) - 0.57721*beta
>>> plt.plot(bins, (1/beta)*np.exp(-(bins - mu)/beta)
... * np.exp(-np.exp(-(bins - mu)/beta)),
... linewidth=2, color='r')
>>> plt.plot(bins, 1/(beta * np.sqrt(2 * np.pi))
... * np.exp(-(bins - mu)**2 / (2 * beta**2)),
... linewidth=2, color='g')
>>> plt.show()
"""
return _mx_nd_np.random.gumbel(loc, scale, size, ctx, out)
def multinomial(n, pvals, size=None, **kwargs):
r"""
Draw samples from a multinomial distribution.
The multinomial distribution is a multivariate generalisation of the binomial distribution.
Take an experiment with one of ``p`` possible outcomes. An example of such an experiment is throwing a dice,
where the outcome can be 1 through 6. Each sample drawn from the distribution represents n such experiments.
Its values, ``X_i = [X_0, X_1, ..., X_p]``, represent the number of times the outcome was ``i``.
Parameters
----------
n : int
Number of experiments.
pvals : sequence of floats, length p
Probabilities of each of the p different outcomes. These should sum to 1.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then ``m * n * k`` samples
are drawn. Default is None, in which case a single value is returned.
Returns
-------
out : ndarray
The drawn samples, of shape size, if that was provided. If not, the shape is ``(N,)``.
In other words, each entry ``out[i,j,...,:]`` is an N-dimensional value drawn from the distribution.
Examples
--------
Throw a dice 1000 times, and 1000 times again:
>>> np.random.multinomial(1000, [1/6.]*6, size=2)
array([[164, 161, 179, 158, 150, 188],
[178, 162, 177, 143, 163, 177]])
A loaded die is more likely to land on number 6:
>>> np.random.multinomial(100, [1/7.]*5 + [2/7.])
array([19, 14, 12, 11, 21, 23])
>>> np.random.multinomial(100, [1.0 / 3, 2.0 / 3])
array([32, 68])
"""
return _mx_nd_np.random.multinomial(n, pvals, size, **kwargs)
# pylint: disable=unused-argument
def multivariate_normal(mean, cov, size=None, check_valid=None, tol=None):
"""
multivariate_normal(mean, cov, size=None, check_valid=None, tol=None)
Draw random samples from a multivariate normal distribution.
The multivariate normal, multinormal or Gaussian distribution is a
generalization of the one-dimensional normal distribution to higher
dimensions. Such a distribution is specified by its mean and
covariance matrix. These parameters are analogous to the mean
(average or "center") and variance (standard deviation, or "width,"
squared) of the one-dimensional normal distribution.
This operator is a little different from the one in official NumPy.
The official NumPy operator only accepts 1-D ndarray as mean and 2-D ndarray as cov,
whereas the operator in MXNet np supports batch operation and auto-broadcasting.
Both `mean` and `cov` may have any number of leading dimensions, which correspond
to a batch shape. They are not necessarily assumed to have the same batch shape,
just ones which can be broadcasted.
Parameters
----------
mean : K-D ndarray, of shape (..., N)
Mean of the N-dimensional distribution.
cov : (K+1)-D ndarray, of shape (..., N, N)
Covariance matrix of the distribution. The last two dimensions must be symmetric and
positive-semidefinite for proper sampling.
size : int or tuple of ints, optional
Given a shape of, for example, ``(m,n,k)``,
``m*n*k`` identically distributed batchs of samples are
generated, and packed in an `m`-by-`n`-by-`k` arrangement.
If no shape is specified, a batch of (`N`-D) sample is returned.
check_valid : { 'warn', 'raise', 'ignore' }, optional
Behavior when the covariance matrix is not positive semidefinite.
(Not supported)
tol : float, optional
Tolerance when checking the singular values in covariance matrix.
cov is cast to double before the check.
(Not supported)
Returns
-------
out : ndarray
The input shape of `mean` and `cov` should satisfy the requirements of broadcasting.
If the parameter `size` is not provided,
the output shape is ``np.broadcast(mean.shape, cov.shape[:-1])``.
Otherwise, the output shape is ``size + np.broadcast(mean.shape, cov.shape[:-1])``
Examples
--------
>>> mean = np.array([1, 2])
>>> cov = np.array([[1, 0], [0, 1]])
>>> x = np.random.multivariate_normal(mean, cov, (3, 3))
>>> x.shape
(3, 3, 2)
The following is probably true, given that 0.6 is roughly twice the
standard deviation:
>>> list((x[0,0,:] - mean) < 0.6)
[True, True] # random
# Performs autobroadcasting when the batch shape of
# `mean` and `cov` is different but compatible.
>>> mean = np.zeros((3,2)) # shape (3, 2)
>>> cov = np.array([[1, 0], [0, 100]]) # shape (2, 2)
>>> x = np.random.multivariate_normal(mean, cov)
>>> x
array([[-1.6115597 , -8.726251 ],
[ 2.2425299 , 2.8104177 ],
[ 0.36229908, -8.386591 ]])
"""
return _mx_nd_np.random.multivariate_normal(mean, cov, size=size, check_valid=None, tol=None)
def choice(a, size=None, replace=True, p=None, ctx=None, out=None):
r"""Generates a random sample from a given 1-D array
Parameters
-----------
a : 1-D array-like or int
If an ndarray, a random sample is generated from its elements.
If an int, the random sample is generated as if a were np.arange(a)
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. Default is None, in which case a
single value is returned.
replace : boolean, optional
Whether the sample is with or without replacement
p : 1-D array-like, optional
The probabilities associated with each entry in a.
If not given the sample assumes a uniform distribution over all
entries in a.
ctx : Context, optional
Device context of output. Default is current context.
Returns
--------
samples : ndarray
The generated random samples
Examples
---------
Generate a uniform random sample from np.arange(5) of size 3:
>>> np.random.choice(5, 3)
array([0, 3, 4])
>>> #This is equivalent to np.random.randint(0,5,3)
Generate a non-uniform random sample from np.arange(5) of size 3:
>>> np.random.choice(5, 3, p=[0.1, 0, 0.3, 0.6, 0])
array([3, 3, 0])
Generate a uniform random sample from np.arange(5) of size 3 without
replacement:
>>> np.random.choice(5, 3, replace=False)
array([3,1,0])
>>> #This is equivalent to np.random.permutation(np.arange(5))[:3]
Generate a non-uniform random sample from np.arange(5) of size
3 without replacement:
>>> np.random.choice(5, 3, replace=False, p=[0.1, 0, 0.3, 0.6, 0])
array([2, 3, 0])
"""
return _mx_nd_np.random.choice(a, size, replace, p, ctx, out)
def rayleigh(scale=1.0, size=None, ctx=None, out=None):
r"""Draw samples from a Rayleigh distribution.
The :math:`\chi` and Weibull distributions are generalizations of the
Rayleigh.
Parameters
----------
scale : float, optional
Scale, also equals the mode. Must be non-negative. Default is 1.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``scale`` is a scalar. Otherwise,
``np.array(scale).size`` samples are drawn.
ctx : Context, optional
Device context of output, default is current context.
out : ``ndarray``, optional
Store output to an existing ``ndarray``.
Returns
-------
out : ndarray or scalar
Drawn samples from the parameterized Rayleigh distribution.
"""
return _mx_nd_np.random.rayleigh(scale, size, ctx, out)
def rand(*size, **kwargs):
r"""Random values in a given shape.
Create an array of the given shape and populate it with random
samples from a uniform distribution over [0, 1).
Parameters
----------
d0, d1, ..., dn : int, optional
The dimensions of the returned array, should be all positive.
If no argument is given a single Python float is returned.
Returns
-------
out : ndarray
Random values.
Examples
--------
>>> np.random.rand(3,2)
array([[ 0.14022471, 0.96360618], #random
[ 0.37601032, 0.25528411], #random
[ 0.49313049, 0.94909878]]) #random
"""
output_shape = ()
for s in size:
output_shape += (s,)
return _mx_nd_np.random.uniform(0, 1, size=output_shape, **kwargs)
def exponential(scale=1.0, size=None, ctx=None, out=None):
r"""Draw samples from an exponential distribution.
Parameters
----------
scale : float or array_like of floats
The scale parameter, :math:`\beta = 1/\lambda`. Must be
non-negative.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``scale`` is a scalar. Otherwise,
``np.array(scale).size`` samples are drawn.
ctx : Context, optional
Device context of output, default is current context.
out : ``ndarray``, optional
Store output to an existing ``ndarray``.
Returns
-------
out : ndarray or scalar
Drawn samples from the parameterized exponential distribution.
"""
return _mx_nd_np.random.exponential(scale, size=size, ctx=ctx, out=out)
def weibull(a, size=None, ctx=None, out=None):
r"""Draw samples from a 1-parameter Weibull distribution with given parameter a
via inversion.
Parameters
----------
a : float or array_like of floats
Shape of the distribution. Must be non-negative.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``a`` is a scalar. Otherwise,
``np.array(a).size`` samples are drawn.
Returns
-------
out : ndarray or scalar
Drawn samples from the 1-parameter Weibull distribution.
Examples
--------
>>> np.random.weibull(a=5)
array(0.9553641)
>>> np.random.weibull(a=5, size=[2,3])
array([[1.0466299 , 1.1320982 , 0.98415005],
[1.1430776 , 0.9532727 , 1.1344457 ]])
>>> np.random.weibull(a=np.array([2,3])
array([0.98843634, 1.0125613 ])
The Weibull distribution is one of a class of Generalized Extreme
Value (GEV) distributions. This class includes the Gumbel and Frechet
distributions.
The probability density for the Weibull distribution is
f(x) = \frac{a}{\lambda}(\frac{x}{\lambda})^{a-1}e^{-(x/\lambda)^a},
where a is the shape and \lambda the scale. The generated 1-parameter Weibull
sample has the scale parameter \lambda = 1.
The Weibull distribution is commonly used in reliability engineering to
model time to failure, in modeling particle sizes, in information retrieval
to model dwell time on pages, in quantitative finance to model risk etc.
"""
return _mx_nd_np.random.weibull(a, size=size, ctx=ctx, out=out)
def pareto(a, size=None, ctx=None, out=None):
r"""Draw samples from a Pareto II or Lomax distribution with specified shape a.
Parameters
----------
a : float or array_like of floats
Shape of the distribution. Must be > 0.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``a`` is a scalar. Otherwise,
``np.array(a).size`` samples are drawn.
Returns
-------
out : ndarray or scalar
Drawn samples from the Pareto distribution.
Examples
--------
>>> np.random.pareto(a=5)
array(0.12749612)
>>> mx.numpy.random.pareto(a=5, size=[2,3])
array([[0.06933999, 0.0344373 , 0.10654891],
[0.0311172 , 0.12911797, 0.03370714]])
>>> np.random.pareto(a=np.array([2,3])
array([0.26636696, 0.15685666])
The probability density for the Pareto distribution is f(x) = \frac{am^a}{x^{a+1}}
where a is the shape and m the scale. Here m is assumed 1. The Pareto distribution
is a power law distribution. Pareto created it to describe the wealth in the economy.
"""
return _mx_nd_np.random.pareto(a, size=size, ctx=ctx, out=out)
def power(a, size=None, ctx=None, out=None):
r"""Draw samples in [0, 1] from a power distribution with given parameter a.
Parameters
----------
a : float or array_like of floats
Shape of the distribution. Must be > 0.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``a`` is a scalar. Otherwise,
``np.array(a).size`` samples are drawn.
Returns
-------
out : ndarray or scalar
Drawn samples from the power distribution.
Examples
--------
>>> np.random.power(a=5)
array(0.8602478)
>>> np.random.power(a=5, size=[2,3])
array([[0.988391 , 0.5153122 , 0.9383134 ],
[0.9078098 , 0.87819266, 0.730635]])
>>> np.random.power(a=np.array([2,3])
array([0.7499419 , 0.88894516])
The probability density function is f(x; a) = ax^{a-1}, 0 \le x \le 1, a>0.
The power distribution is just the inverse of the Pareto distribution and
a special case of the Beta distribution.
"""
return _mx_nd_np.random.power(a, size=size, ctx=ctx, out=out)
def shuffle(x):
"""
Modify a sequence in-place by shuffling its contents.
This function only shuffles the array along the first axis of a
multi-dimensional array. The order of sub-arrays is changed but
their contents remain the same.
Parameters
----------
x: ndarray
The array or list to be shuffled.
Examples
--------
>>> arr = np.arange(10)
>>> np.random.shuffle(arr)
>>> arr
array([5., 1., 0., 6., 7., 3., 9., 8., 4., 2.]) # random
Multi-dimensional arrays are only shuffled along the first axis:
>>> arr = np.arange(9).reshape((3, 3))
>>> np.random.shuffle(arr)
>>> arr
array([[6., 7., 8.], # random
[3., 4., 5.],
[0., 1., 2.]])
"""
_mx_nd_np.random.shuffle(x)
def gamma(shape, scale=1.0, size=None, dtype=None, ctx=None, out=None):
"""Draw samples from a Gamma distribution.
Samples are drawn from a Gamma distribution with specified parameters,
`shape` (sometimes designated "k") and `scale` (sometimes designated
"theta"), where both parameters are > 0.
The Gamma distribution is often used to model the times to failure of
electronic components, and arises naturally in processes for which the
waiting times between Poisson distributed events are relevant.
Parameters
----------
shape : float or array_like of floats
The shape of the gamma distribution. Should be greater than zero.
scale : float or array_like of floats, optional
The scale of the gamma distribution. Should be greater than zero.
Default is equal to 1.
dtype : {'float16', 'float32', 'float64'}, optional
Data type of output samples. Default is 'float32'.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``shape`` and ``scale`` are both scalars.
Otherwise, ``np.broadcast(shape, scale).size`` samples are drawn.
ctx : Context, optional
Device context of output. Default is current context.
Returns
-------
out : ndarray or scalar
Drawn samples from the parameterized gamma distribution.
"""
return _mx_nd_np.random.gamma(shape, scale, size, dtype, ctx, out)
def beta(a, b, size=None, dtype=None, ctx=None):
r"""Draw samples from a Beta distribution.
The Beta distribution is a special case of the Dirichlet distribution,
and is related to the Gamma distribution. It has the probability
distribution function
.. math:: f(x; a,b) = \frac{1}{B(\alpha, \beta)} x^{\alpha - 1}
(1 - x)^{\beta - 1},
where the normalisation, B, is the beta function,
.. math:: B(\alpha, \beta) = \int_0^1 t^{\alpha - 1}
(1 - t)^{\beta - 1} dt.
It is often seen in Bayesian inference and order statistics.
Parameters
----------
a : float or array_like of floats
Alpha, positive (>0).
b : float or array_like of floats
Beta, positive (>0).
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``a`` and ``b`` are both scalars.
Otherwise, ``np.broadcast(a, b).size`` samples are drawn.
dtype : {'float16', 'float32', 'float64'}, optional
Data type of output samples. Default is 'float32'.
Dtype 'float32' or 'float64' is strongly recommended,
since lower precision might lead to out of range issue.
ctx : Context, optional
Device context of output. Default is current context.
Notes
-----
To use this operator with scalars as input, please run
``npx.set_np()`` first.
Returns
-------
out : ndarray or scalar
Drawn samples from the parameterized beta distribution.
"""
return _mx_nd_np.random.beta(a, b, size=size, dtype=dtype, ctx=ctx)
def f(dfnum, dfden, size=None, ctx=None):
r"""Draw samples from an F distribution.
Samples are drawn from an F distribution with specified parameters,
`dfnum` (degrees of freedom in numerator) and `dfden` (degrees of
freedom in denominator), where both parameters must be greater than
zero.
The random variate of the F distribution (also known as the
Fisher distribution) is a continuous probability distribution
that arises in ANOVA tests, and is the ratio of two chi-square
variates.
Parameters
----------
dfnum : float or ndarray of floats
Degrees of freedom in numerator, must be > 0.
dfden : float or ndarray of float
Degrees of freedom in denominator, must be > 0.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``dfnum`` and ``dfden`` are both scalars.
Otherwise, ``np.broadcast(dfnum, dfden).size`` samples are drawn.
ctx : Context, optional
Device context of output. Default is current context.
Returns
-------
out : ndarray or scalar
Drawn samples from the parameterized Fisher distribution.
Examples
--------
An example from Glantz[1], pp 47-40:
Two groups, children of diabetics (25 people) and children from people
without diabetes (25 controls). Fasting blood glucose was measured,
case group had a mean value of 86.1, controls had a mean value of
82.2. Standard deviations were 2.09 and 2.49 respectively. Are these
data consistent with the null hypothesis that the parents diabetic
status does not affect their children's blood glucose levels?
Calculating the F statistic from the data gives a value of 36.01.
Draw samples from the distribution:
>>> dfnum = 1. # between group degrees of freedom
>>> dfden = 48. # within groups degrees of freedom
>>> s = np.random.f(dfnum, dfden, 1000)
The lower bound for the top 1% of the samples is :
>>> np.sort(s)[-10]
7.61988120985 # random
So there is about a 1% chance that the F statistic will exceed 7.62,
the measured value is 36, so the null hypothesis is rejected at the 1%
level.
"""
return _mx_nd_np.random.f(dfnum, dfden, size=size, ctx=ctx)
def chisquare(df, size=None, dtype=None, ctx=None):
r"""Draw samples from a chi-square distribution.
When `df` independent random variables, each with standard normal
distributions (mean 0, variance 1), are squared and summed, the
resulting distribution is chi-square (see Notes). This distribution
is often used in hypothesis testing.
Parameters
----------
df : float or ndarray of floats
Number of degrees of freedom, must be > 0.
size : int or tuple of ints, optional
Output shape. If the given shape is, e.g., ``(m, n, k)``, then
``m * n * k`` samples are drawn. If size is ``None`` (default),
a single value is returned if ``df`` is a scalar. Otherwise,
``np.array(df).size`` samples are drawn.
dtype : {'float16', 'float32', 'float64'}, optional
Data type of output samples. Default is 'float32'.
ctx : Context, optional
Device context of output. Default is current context.
Returns
-------
out : ndarray or scalar
Drawn samples from the parameterized `chi-square distribution` [1]_.
Raises
------
ValueError
When `df` <= 0 or when an inappropriate `size`
is given.
Notes
-----
The variable obtained by summing the squares of `df` independent,
standard normally distributed random variables:
.. math:: Q = \sum_{i=0}^{\mathtt{df}} X^2_i
is chi-square distributed, denoted
.. math:: Q \sim \chi^2_k.
The probability density function of the chi-squared distribution is
.. math:: p(x) = \frac{(1/2)^{k/2}}{\Gamma(k/2)}
x^{k/2 - 1} e^{-x/2},
where :math:`\Gamma` is the gamma function,
.. math:: \Gamma(x) = \int_0^{-\infty} t^{x - 1} e^{-t} dt.
References
----------
.. [1] NIST "Engineering Statistics Handbook"
https://www.itl.nist.gov/div898/handbook/eda/section3/eda3666.htm
Examples
--------
>>> np.random.chisquare(2,4)
array([ 1.89920014, 9.00867716, 3.13710533, 5.62318272]) # random
"""
return _mx_nd_np.random.chisquare(df, size=size, dtype=dtype, ctx=ctx)
def randn(*size, **kwargs):
r"""Return a sample (or samples) from the "standard normal" distribution.
If positive, int_like or int-convertible arguments are provided,
`randn` generates an array of shape ``(d0, d1, ..., dn)``, filled
with random floats sampled from a univariate "normal" (Gaussian)
distribution of mean 0 and variance 1 (if any of the :math:`d_i` are
floats, they are first converted to integers by truncation). A single
float randomly sampled from the distribution is returned if no
argument is provided.
This is a convenience function. If you want an interface that takes a
tuple as the first argument, use `numpy.random.standard_normal` instead.
Parameters
----------
d0, d1, ..., dn : int, optional
The dimensions of the returned array, should be all positive.
If no argument is given a single Python float is returned.
Returns
-------
Z : ndarray
A ``(d0, d1, ..., dn)``-shaped array of floating-point samples from
the standard normal distribution, or a single such float if
no parameters were supplied.
Notes
-----
For random samples from :math:`N(\mu, \sigma^2)`, use:
``sigma * np.random.randn(...) + mu``
Examples
--------
>>> np.random.randn()
2.1923875335537315 #random
Two-by-four array of samples from N(3, 6.25):
>>> 2.5 * np.random.randn(2, 4) + 3
array([[-4.49401501, 4.00950034, -1.81814867, 7.29718677], #random
[ 0.39924804, 4.68456316, 4.99394529, 4.84057254]]) #random
"""
output_shape = ()
for s in size:
output_shape += (s,)
return _mx_nd_np.random.normal(0, 1, size=output_shape, **kwargs)
def laplace(loc=0.0, scale=1.0, size=None, dtype=None, ctx=None, out=None):
r"""Draw random samples from a Laplace distribution.
Samples are distributed according to a Laplace distribution parametrized
by *loc* (mean) and *scale* (the exponential decay).
Parameters
----------
loc : float, The position of the distribution peak.
scale : float, the exponential decay.
size : int or tuple of ints, optional. Output shape.
If the given shape is, e.g., (m, n, k), then m * n * k samples are drawn.
Default is None, in which case a single value is returned.
dtype : {'float16', 'float32', 'float64'}, optional
Data type of output samples. Default is 'float32'
ctx : Context, optional
Device context of output. Default is current context.
out : ``ndarray``, optional
Store output to an existing ``ndarray``.
Returns
-------
out : ndarray
Drawn samples from the parameterized Laplace distribution.
"""
return _mx_nd_np.random.laplace(loc, scale, size, dtype, ctx, out)
| apache-2.0 |
ammarkhann/FinalSeniorCode | lib/python2.7/site-packages/pandas/tests/frame/test_query_eval.py | 11 | 42389 | # -*- coding: utf-8 -*-
from __future__ import print_function
import operator
import pytest
from pandas.compat import (zip, range, lrange, StringIO)
from pandas import DataFrame, Series, Index, MultiIndex, date_range
import pandas as pd
import numpy as np
from numpy.random import randn
from pandas.util.testing import (assert_series_equal,
assert_frame_equal,
makeCustomDataframe as mkdf)
import pandas.util.testing as tm
from pandas.core.computation import _NUMEXPR_INSTALLED
from pandas.tests.frame.common import TestData
PARSERS = 'python', 'pandas'
ENGINES = 'python', 'numexpr'
@pytest.fixture(params=PARSERS, ids=lambda x: x)
def parser(request):
return request.param
@pytest.fixture(params=ENGINES, ids=lambda x: x)
def engine(request):
return request.param
def skip_if_no_pandas_parser(parser):
if parser != 'pandas':
pytest.skip("cannot evaluate with parser {0!r}".format(parser))
def skip_if_no_ne(engine='numexpr'):
if engine == 'numexpr':
if not _NUMEXPR_INSTALLED:
pytest.skip("cannot query engine numexpr when numexpr not "
"installed")
class TestCompat(object):
def setup_method(self, method):
self.df = DataFrame({'A': [1, 2, 3]})
self.expected1 = self.df[self.df.A > 0]
self.expected2 = self.df.A + 1
def test_query_default(self):
# GH 12749
# this should always work, whether _NUMEXPR_INSTALLED or not
df = self.df
result = df.query('A>0')
assert_frame_equal(result, self.expected1)
result = df.eval('A+1')
assert_series_equal(result, self.expected2, check_names=False)
def test_query_None(self):
df = self.df
result = df.query('A>0', engine=None)
assert_frame_equal(result, self.expected1)
result = df.eval('A+1', engine=None)
assert_series_equal(result, self.expected2, check_names=False)
def test_query_python(self):
df = self.df
result = df.query('A>0', engine='python')
assert_frame_equal(result, self.expected1)
result = df.eval('A+1', engine='python')
assert_series_equal(result, self.expected2, check_names=False)
def test_query_numexpr(self):
df = self.df
if _NUMEXPR_INSTALLED:
result = df.query('A>0', engine='numexpr')
assert_frame_equal(result, self.expected1)
result = df.eval('A+1', engine='numexpr')
assert_series_equal(result, self.expected2, check_names=False)
else:
pytest.raises(ImportError,
lambda: df.query('A>0', engine='numexpr'))
pytest.raises(ImportError,
lambda: df.eval('A+1', engine='numexpr'))
class TestDataFrameEval(TestData):
def test_ops(self):
# tst ops and reversed ops in evaluation
# GH7198
# smaller hits python, larger hits numexpr
for n in [4, 4000]:
df = DataFrame(1, index=range(n), columns=list('abcd'))
df.iloc[0] = 2
m = df.mean()
for op_str, op, rop in [('+', '__add__', '__radd__'),
('-', '__sub__', '__rsub__'),
('*', '__mul__', '__rmul__'),
('/', '__truediv__', '__rtruediv__')]:
base = (DataFrame(np.tile(m.values, n) # noqa
.reshape(n, -1),
columns=list('abcd')))
expected = eval("base{op}df".format(op=op_str))
# ops as strings
result = eval("m{op}df".format(op=op_str))
assert_frame_equal(result, expected)
# these are commutative
if op in ['+', '*']:
result = getattr(df, op)(m)
assert_frame_equal(result, expected)
# these are not
elif op in ['-', '/']:
result = getattr(df, rop)(m)
assert_frame_equal(result, expected)
# GH7192
df = DataFrame(dict(A=np.random.randn(25000)))
df.iloc[0:5] = np.nan
expected = (1 - np.isnan(df.iloc[0:25]))
result = (1 - np.isnan(df)).iloc[0:25]
assert_frame_equal(result, expected)
def test_query_non_str(self):
# GH 11485
df = pd.DataFrame({'A': [1, 2, 3], 'B': ['a', 'b', 'b']})
msg = "expr must be a string to be evaluated"
with tm.assert_raises_regex(ValueError, msg):
df.query(lambda x: x.B == "b")
with tm.assert_raises_regex(ValueError, msg):
df.query(111)
def test_query_empty_string(self):
# GH 13139
df = pd.DataFrame({'A': [1, 2, 3]})
msg = "expr cannot be an empty string"
with tm.assert_raises_regex(ValueError, msg):
df.query('')
def test_eval_resolvers_as_list(self):
# GH 14095
df = DataFrame(randn(10, 2), columns=list('ab'))
dict1 = {'a': 1}
dict2 = {'b': 2}
assert (df.eval('a + b', resolvers=[dict1, dict2]) ==
dict1['a'] + dict2['b'])
assert (pd.eval('a + b', resolvers=[dict1, dict2]) ==
dict1['a'] + dict2['b'])
class TestDataFrameQueryWithMultiIndex(object):
def test_query_with_named_multiindex(self, parser, engine):
tm.skip_if_no_ne(engine)
skip_if_no_pandas_parser(parser)
a = np.random.choice(['red', 'green'], size=10)
b = np.random.choice(['eggs', 'ham'], size=10)
index = MultiIndex.from_arrays([a, b], names=['color', 'food'])
df = DataFrame(randn(10, 2), index=index)
ind = Series(df.index.get_level_values('color').values, index=index,
name='color')
# equality
res1 = df.query('color == "red"', parser=parser, engine=engine)
res2 = df.query('"red" == color', parser=parser, engine=engine)
exp = df[ind == 'red']
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
# inequality
res1 = df.query('color != "red"', parser=parser, engine=engine)
res2 = df.query('"red" != color', parser=parser, engine=engine)
exp = df[ind != 'red']
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
# list equality (really just set membership)
res1 = df.query('color == ["red"]', parser=parser, engine=engine)
res2 = df.query('["red"] == color', parser=parser, engine=engine)
exp = df[ind.isin(['red'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
res1 = df.query('color != ["red"]', parser=parser, engine=engine)
res2 = df.query('["red"] != color', parser=parser, engine=engine)
exp = df[~ind.isin(['red'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
# in/not in ops
res1 = df.query('["red"] in color', parser=parser, engine=engine)
res2 = df.query('"red" in color', parser=parser, engine=engine)
exp = df[ind.isin(['red'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
res1 = df.query('["red"] not in color', parser=parser, engine=engine)
res2 = df.query('"red" not in color', parser=parser, engine=engine)
exp = df[~ind.isin(['red'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
def test_query_with_unnamed_multiindex(self, parser, engine):
tm.skip_if_no_ne(engine)
skip_if_no_pandas_parser(parser)
a = np.random.choice(['red', 'green'], size=10)
b = np.random.choice(['eggs', 'ham'], size=10)
index = MultiIndex.from_arrays([a, b])
df = DataFrame(randn(10, 2), index=index)
ind = Series(df.index.get_level_values(0).values, index=index)
res1 = df.query('ilevel_0 == "red"', parser=parser, engine=engine)
res2 = df.query('"red" == ilevel_0', parser=parser, engine=engine)
exp = df[ind == 'red']
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
# inequality
res1 = df.query('ilevel_0 != "red"', parser=parser, engine=engine)
res2 = df.query('"red" != ilevel_0', parser=parser, engine=engine)
exp = df[ind != 'red']
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
# list equality (really just set membership)
res1 = df.query('ilevel_0 == ["red"]', parser=parser, engine=engine)
res2 = df.query('["red"] == ilevel_0', parser=parser, engine=engine)
exp = df[ind.isin(['red'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
res1 = df.query('ilevel_0 != ["red"]', parser=parser, engine=engine)
res2 = df.query('["red"] != ilevel_0', parser=parser, engine=engine)
exp = df[~ind.isin(['red'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
# in/not in ops
res1 = df.query('["red"] in ilevel_0', parser=parser, engine=engine)
res2 = df.query('"red" in ilevel_0', parser=parser, engine=engine)
exp = df[ind.isin(['red'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
res1 = df.query('["red"] not in ilevel_0', parser=parser,
engine=engine)
res2 = df.query('"red" not in ilevel_0', parser=parser, engine=engine)
exp = df[~ind.isin(['red'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
# ## LEVEL 1
ind = Series(df.index.get_level_values(1).values, index=index)
res1 = df.query('ilevel_1 == "eggs"', parser=parser, engine=engine)
res2 = df.query('"eggs" == ilevel_1', parser=parser, engine=engine)
exp = df[ind == 'eggs']
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
# inequality
res1 = df.query('ilevel_1 != "eggs"', parser=parser, engine=engine)
res2 = df.query('"eggs" != ilevel_1', parser=parser, engine=engine)
exp = df[ind != 'eggs']
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
# list equality (really just set membership)
res1 = df.query('ilevel_1 == ["eggs"]', parser=parser, engine=engine)
res2 = df.query('["eggs"] == ilevel_1', parser=parser, engine=engine)
exp = df[ind.isin(['eggs'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
res1 = df.query('ilevel_1 != ["eggs"]', parser=parser, engine=engine)
res2 = df.query('["eggs"] != ilevel_1', parser=parser, engine=engine)
exp = df[~ind.isin(['eggs'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
# in/not in ops
res1 = df.query('["eggs"] in ilevel_1', parser=parser, engine=engine)
res2 = df.query('"eggs" in ilevel_1', parser=parser, engine=engine)
exp = df[ind.isin(['eggs'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
res1 = df.query('["eggs"] not in ilevel_1', parser=parser,
engine=engine)
res2 = df.query('"eggs" not in ilevel_1', parser=parser, engine=engine)
exp = df[~ind.isin(['eggs'])]
assert_frame_equal(res1, exp)
assert_frame_equal(res2, exp)
def test_query_with_partially_named_multiindex(self, parser, engine):
tm.skip_if_no_ne(engine)
skip_if_no_pandas_parser(parser)
a = np.random.choice(['red', 'green'], size=10)
b = np.arange(10)
index = MultiIndex.from_arrays([a, b])
index.names = [None, 'rating']
df = DataFrame(randn(10, 2), index=index)
res = df.query('rating == 1', parser=parser, engine=engine)
ind = Series(df.index.get_level_values('rating').values, index=index,
name='rating')
exp = df[ind == 1]
assert_frame_equal(res, exp)
res = df.query('rating != 1', parser=parser, engine=engine)
ind = Series(df.index.get_level_values('rating').values, index=index,
name='rating')
exp = df[ind != 1]
assert_frame_equal(res, exp)
res = df.query('ilevel_0 == "red"', parser=parser, engine=engine)
ind = Series(df.index.get_level_values(0).values, index=index)
exp = df[ind == "red"]
assert_frame_equal(res, exp)
res = df.query('ilevel_0 != "red"', parser=parser, engine=engine)
ind = Series(df.index.get_level_values(0).values, index=index)
exp = df[ind != "red"]
assert_frame_equal(res, exp)
def test_query_multiindex_get_index_resolvers(self):
df = mkdf(10, 3, r_idx_nlevels=2, r_idx_names=['spam', 'eggs'])
resolvers = df._get_index_resolvers()
def to_series(mi, level):
level_values = mi.get_level_values(level)
s = level_values.to_series()
s.index = mi
return s
col_series = df.columns.to_series()
expected = {'index': df.index,
'columns': col_series,
'spam': to_series(df.index, 'spam'),
'eggs': to_series(df.index, 'eggs'),
'C0': col_series}
for k, v in resolvers.items():
if isinstance(v, Index):
assert v.is_(expected[k])
elif isinstance(v, Series):
assert_series_equal(v, expected[k])
else:
raise AssertionError("object must be a Series or Index")
def test_raise_on_panel_with_multiindex(self, parser, engine):
tm.skip_if_no_ne()
p = tm.makePanel(7)
p.items = tm.makeCustomIndex(len(p.items), nlevels=2)
with pytest.raises(NotImplementedError):
pd.eval('p + 1', parser=parser, engine=engine)
def test_raise_on_panel4d_with_multiindex(self, parser, engine):
tm.skip_if_no_ne()
p4d = tm.makePanel4D(7)
p4d.items = tm.makeCustomIndex(len(p4d.items), nlevels=2)
with pytest.raises(NotImplementedError):
pd.eval('p4d + 1', parser=parser, engine=engine)
class TestDataFrameQueryNumExprPandas(object):
@classmethod
def setup_class(cls):
cls.engine = 'numexpr'
cls.parser = 'pandas'
tm.skip_if_no_ne(cls.engine)
@classmethod
def teardown_class(cls):
del cls.engine, cls.parser
def test_date_query_with_attribute_access(self):
engine, parser = self.engine, self.parser
skip_if_no_pandas_parser(parser)
df = DataFrame(randn(5, 3))
df['dates1'] = date_range('1/1/2012', periods=5)
df['dates2'] = date_range('1/1/2013', periods=5)
df['dates3'] = date_range('1/1/2014', periods=5)
res = df.query('@df.dates1 < 20130101 < @df.dates3', engine=engine,
parser=parser)
expec = df[(df.dates1 < '20130101') & ('20130101' < df.dates3)]
assert_frame_equal(res, expec)
def test_date_query_no_attribute_access(self):
engine, parser = self.engine, self.parser
df = DataFrame(randn(5, 3))
df['dates1'] = date_range('1/1/2012', periods=5)
df['dates2'] = date_range('1/1/2013', periods=5)
df['dates3'] = date_range('1/1/2014', periods=5)
res = df.query('dates1 < 20130101 < dates3', engine=engine,
parser=parser)
expec = df[(df.dates1 < '20130101') & ('20130101' < df.dates3)]
assert_frame_equal(res, expec)
def test_date_query_with_NaT(self):
engine, parser = self.engine, self.parser
n = 10
df = DataFrame(randn(n, 3))
df['dates1'] = date_range('1/1/2012', periods=n)
df['dates2'] = date_range('1/1/2013', periods=n)
df['dates3'] = date_range('1/1/2014', periods=n)
df.loc[np.random.rand(n) > 0.5, 'dates1'] = pd.NaT
df.loc[np.random.rand(n) > 0.5, 'dates3'] = pd.NaT
res = df.query('dates1 < 20130101 < dates3', engine=engine,
parser=parser)
expec = df[(df.dates1 < '20130101') & ('20130101' < df.dates3)]
assert_frame_equal(res, expec)
def test_date_index_query(self):
engine, parser = self.engine, self.parser
n = 10
df = DataFrame(randn(n, 3))
df['dates1'] = date_range('1/1/2012', periods=n)
df['dates3'] = date_range('1/1/2014', periods=n)
df.set_index('dates1', inplace=True, drop=True)
res = df.query('index < 20130101 < dates3', engine=engine,
parser=parser)
expec = df[(df.index < '20130101') & ('20130101' < df.dates3)]
assert_frame_equal(res, expec)
def test_date_index_query_with_NaT(self):
engine, parser = self.engine, self.parser
n = 10
df = DataFrame(randn(n, 3))
df['dates1'] = date_range('1/1/2012', periods=n)
df['dates3'] = date_range('1/1/2014', periods=n)
df.iloc[0, 0] = pd.NaT
df.set_index('dates1', inplace=True, drop=True)
res = df.query('index < 20130101 < dates3', engine=engine,
parser=parser)
expec = df[(df.index < '20130101') & ('20130101' < df.dates3)]
assert_frame_equal(res, expec)
def test_date_index_query_with_NaT_duplicates(self):
engine, parser = self.engine, self.parser
n = 10
d = {}
d['dates1'] = date_range('1/1/2012', periods=n)
d['dates3'] = date_range('1/1/2014', periods=n)
df = DataFrame(d)
df.loc[np.random.rand(n) > 0.5, 'dates1'] = pd.NaT
df.set_index('dates1', inplace=True, drop=True)
res = df.query('dates1 < 20130101 < dates3', engine=engine,
parser=parser)
expec = df[(df.index.to_series() < '20130101') &
('20130101' < df.dates3)]
assert_frame_equal(res, expec)
def test_date_query_with_non_date(self):
engine, parser = self.engine, self.parser
n = 10
df = DataFrame({'dates': date_range('1/1/2012', periods=n),
'nondate': np.arange(n)})
ops = '==', '!=', '<', '>', '<=', '>='
for op in ops:
with pytest.raises(TypeError):
df.query('dates %s nondate' % op, parser=parser, engine=engine)
def test_query_syntax_error(self):
engine, parser = self.engine, self.parser
df = DataFrame({"i": lrange(10), "+": lrange(3, 13),
"r": lrange(4, 14)})
with pytest.raises(SyntaxError):
df.query('i - +', engine=engine, parser=parser)
def test_query_scope(self):
from pandas.core.computation.ops import UndefinedVariableError
engine, parser = self.engine, self.parser
skip_if_no_pandas_parser(parser)
df = DataFrame(np.random.randn(20, 2), columns=list('ab'))
a, b = 1, 2 # noqa
res = df.query('a > b', engine=engine, parser=parser)
expected = df[df.a > df.b]
assert_frame_equal(res, expected)
res = df.query('@a > b', engine=engine, parser=parser)
expected = df[a > df.b]
assert_frame_equal(res, expected)
# no local variable c
with pytest.raises(UndefinedVariableError):
df.query('@a > b > @c', engine=engine, parser=parser)
# no column named 'c'
with pytest.raises(UndefinedVariableError):
df.query('@a > b > c', engine=engine, parser=parser)
def test_query_doesnt_pickup_local(self):
from pandas.core.computation.ops import UndefinedVariableError
engine, parser = self.engine, self.parser
n = m = 10
df = DataFrame(np.random.randint(m, size=(n, 3)), columns=list('abc'))
# we don't pick up the local 'sin'
with pytest.raises(UndefinedVariableError):
df.query('sin > 5', engine=engine, parser=parser)
def test_query_builtin(self):
from pandas.core.computation.engines import NumExprClobberingError
engine, parser = self.engine, self.parser
n = m = 10
df = DataFrame(np.random.randint(m, size=(n, 3)), columns=list('abc'))
df.index.name = 'sin'
with tm.assert_raises_regex(NumExprClobberingError,
'Variables in expression.+'):
df.query('sin > 5', engine=engine, parser=parser)
def test_query(self):
engine, parser = self.engine, self.parser
df = DataFrame(np.random.randn(10, 3), columns=['a', 'b', 'c'])
assert_frame_equal(df.query('a < b', engine=engine, parser=parser),
df[df.a < df.b])
assert_frame_equal(df.query('a + b > b * c', engine=engine,
parser=parser),
df[df.a + df.b > df.b * df.c])
def test_query_index_with_name(self):
engine, parser = self.engine, self.parser
df = DataFrame(np.random.randint(10, size=(10, 3)),
index=Index(range(10), name='blob'),
columns=['a', 'b', 'c'])
res = df.query('(blob < 5) & (a < b)', engine=engine, parser=parser)
expec = df[(df.index < 5) & (df.a < df.b)]
assert_frame_equal(res, expec)
res = df.query('blob < b', engine=engine, parser=parser)
expec = df[df.index < df.b]
assert_frame_equal(res, expec)
def test_query_index_without_name(self):
engine, parser = self.engine, self.parser
df = DataFrame(np.random.randint(10, size=(10, 3)),
index=range(10), columns=['a', 'b', 'c'])
# "index" should refer to the index
res = df.query('index < b', engine=engine, parser=parser)
expec = df[df.index < df.b]
assert_frame_equal(res, expec)
# test against a scalar
res = df.query('index < 5', engine=engine, parser=parser)
expec = df[df.index < 5]
assert_frame_equal(res, expec)
def test_nested_scope(self):
engine = self.engine
parser = self.parser
skip_if_no_pandas_parser(parser)
df = DataFrame(np.random.randn(5, 3))
df2 = DataFrame(np.random.randn(5, 3))
expected = df[(df > 0) & (df2 > 0)]
result = df.query('(@df > 0) & (@df2 > 0)', engine=engine,
parser=parser)
assert_frame_equal(result, expected)
result = pd.eval('df[df > 0 and df2 > 0]', engine=engine,
parser=parser)
assert_frame_equal(result, expected)
result = pd.eval('df[df > 0 and df2 > 0 and df[df > 0] > 0]',
engine=engine, parser=parser)
expected = df[(df > 0) & (df2 > 0) & (df[df > 0] > 0)]
assert_frame_equal(result, expected)
result = pd.eval('df[(df>0) & (df2>0)]', engine=engine, parser=parser)
expected = df.query('(@df>0) & (@df2>0)', engine=engine, parser=parser)
assert_frame_equal(result, expected)
def test_nested_raises_on_local_self_reference(self):
from pandas.core.computation.ops import UndefinedVariableError
df = DataFrame(np.random.randn(5, 3))
# can't reference ourself b/c we're a local so @ is necessary
with pytest.raises(UndefinedVariableError):
df.query('df > 0', engine=self.engine, parser=self.parser)
def test_local_syntax(self):
skip_if_no_pandas_parser(self.parser)
engine, parser = self.engine, self.parser
df = DataFrame(randn(100, 10), columns=list('abcdefghij'))
b = 1
expect = df[df.a < b]
result = df.query('a < @b', engine=engine, parser=parser)
assert_frame_equal(result, expect)
expect = df[df.a < df.b]
result = df.query('a < b', engine=engine, parser=parser)
assert_frame_equal(result, expect)
def test_chained_cmp_and_in(self):
skip_if_no_pandas_parser(self.parser)
engine, parser = self.engine, self.parser
cols = list('abc')
df = DataFrame(randn(100, len(cols)), columns=cols)
res = df.query('a < b < c and a not in b not in c', engine=engine,
parser=parser)
ind = (df.a < df.b) & (df.b < df.c) & ~df.b.isin(df.a) & ~df.c.isin(df.b) # noqa
expec = df[ind]
assert_frame_equal(res, expec)
def test_local_variable_with_in(self):
engine, parser = self.engine, self.parser
skip_if_no_pandas_parser(parser)
a = Series(np.random.randint(3, size=15), name='a')
b = Series(np.random.randint(10, size=15), name='b')
df = DataFrame({'a': a, 'b': b})
expected = df.loc[(df.b - 1).isin(a)]
result = df.query('b - 1 in a', engine=engine, parser=parser)
assert_frame_equal(expected, result)
b = Series(np.random.randint(10, size=15), name='b')
expected = df.loc[(b - 1).isin(a)]
result = df.query('@b - 1 in a', engine=engine, parser=parser)
assert_frame_equal(expected, result)
def test_at_inside_string(self):
engine, parser = self.engine, self.parser
skip_if_no_pandas_parser(parser)
c = 1 # noqa
df = DataFrame({'a': ['a', 'a', 'b', 'b', '@c', '@c']})
result = df.query('a == "@c"', engine=engine, parser=parser)
expected = df[df.a == "@c"]
assert_frame_equal(result, expected)
def test_query_undefined_local(self):
from pandas.core.computation.ops import UndefinedVariableError
engine, parser = self.engine, self.parser
skip_if_no_pandas_parser(parser)
df = DataFrame(np.random.rand(10, 2), columns=list('ab'))
with tm.assert_raises_regex(UndefinedVariableError,
"local variable 'c' is not defined"):
df.query('a == @c', engine=engine, parser=parser)
def test_index_resolvers_come_after_columns_with_the_same_name(self):
n = 1 # noqa
a = np.r_[20:101:20]
df = DataFrame({'index': a, 'b': np.random.randn(a.size)})
df.index.name = 'index'
result = df.query('index > 5', engine=self.engine, parser=self.parser)
expected = df[df['index'] > 5]
assert_frame_equal(result, expected)
df = DataFrame({'index': a,
'b': np.random.randn(a.size)})
result = df.query('ilevel_0 > 5', engine=self.engine,
parser=self.parser)
expected = df.loc[df.index[df.index > 5]]
assert_frame_equal(result, expected)
df = DataFrame({'a': a, 'b': np.random.randn(a.size)})
df.index.name = 'a'
result = df.query('a > 5', engine=self.engine, parser=self.parser)
expected = df[df.a > 5]
assert_frame_equal(result, expected)
result = df.query('index > 5', engine=self.engine, parser=self.parser)
expected = df.loc[df.index[df.index > 5]]
assert_frame_equal(result, expected)
def test_inf(self):
n = 10
df = DataFrame({'a': np.random.rand(n), 'b': np.random.rand(n)})
df.loc[::2, 0] = np.inf
ops = '==', '!='
d = dict(zip(ops, (operator.eq, operator.ne)))
for op, f in d.items():
q = 'a %s inf' % op
expected = df[f(df.a, np.inf)]
result = df.query(q, engine=self.engine, parser=self.parser)
assert_frame_equal(result, expected)
class TestDataFrameQueryNumExprPython(TestDataFrameQueryNumExprPandas):
@classmethod
def setup_class(cls):
super(TestDataFrameQueryNumExprPython, cls).setup_class()
cls.engine = 'numexpr'
cls.parser = 'python'
tm.skip_if_no_ne(cls.engine)
cls.frame = TestData().frame
def test_date_query_no_attribute_access(self):
engine, parser = self.engine, self.parser
df = DataFrame(randn(5, 3))
df['dates1'] = date_range('1/1/2012', periods=5)
df['dates2'] = date_range('1/1/2013', periods=5)
df['dates3'] = date_range('1/1/2014', periods=5)
res = df.query('(dates1 < 20130101) & (20130101 < dates3)',
engine=engine, parser=parser)
expec = df[(df.dates1 < '20130101') & ('20130101' < df.dates3)]
assert_frame_equal(res, expec)
def test_date_query_with_NaT(self):
engine, parser = self.engine, self.parser
n = 10
df = DataFrame(randn(n, 3))
df['dates1'] = date_range('1/1/2012', periods=n)
df['dates2'] = date_range('1/1/2013', periods=n)
df['dates3'] = date_range('1/1/2014', periods=n)
df.loc[np.random.rand(n) > 0.5, 'dates1'] = pd.NaT
df.loc[np.random.rand(n) > 0.5, 'dates3'] = pd.NaT
res = df.query('(dates1 < 20130101) & (20130101 < dates3)',
engine=engine, parser=parser)
expec = df[(df.dates1 < '20130101') & ('20130101' < df.dates3)]
assert_frame_equal(res, expec)
def test_date_index_query(self):
engine, parser = self.engine, self.parser
n = 10
df = DataFrame(randn(n, 3))
df['dates1'] = date_range('1/1/2012', periods=n)
df['dates3'] = date_range('1/1/2014', periods=n)
df.set_index('dates1', inplace=True, drop=True)
res = df.query('(index < 20130101) & (20130101 < dates3)',
engine=engine, parser=parser)
expec = df[(df.index < '20130101') & ('20130101' < df.dates3)]
assert_frame_equal(res, expec)
def test_date_index_query_with_NaT(self):
engine, parser = self.engine, self.parser
n = 10
df = DataFrame(randn(n, 3))
df['dates1'] = date_range('1/1/2012', periods=n)
df['dates3'] = date_range('1/1/2014', periods=n)
df.iloc[0, 0] = pd.NaT
df.set_index('dates1', inplace=True, drop=True)
res = df.query('(index < 20130101) & (20130101 < dates3)',
engine=engine, parser=parser)
expec = df[(df.index < '20130101') & ('20130101' < df.dates3)]
assert_frame_equal(res, expec)
def test_date_index_query_with_NaT_duplicates(self):
engine, parser = self.engine, self.parser
n = 10
df = DataFrame(randn(n, 3))
df['dates1'] = date_range('1/1/2012', periods=n)
df['dates3'] = date_range('1/1/2014', periods=n)
df.loc[np.random.rand(n) > 0.5, 'dates1'] = pd.NaT
df.set_index('dates1', inplace=True, drop=True)
with pytest.raises(NotImplementedError):
df.query('index < 20130101 < dates3', engine=engine, parser=parser)
def test_nested_scope(self):
from pandas.core.computation.ops import UndefinedVariableError
engine = self.engine
parser = self.parser
# smoke test
x = 1 # noqa
result = pd.eval('x + 1', engine=engine, parser=parser)
assert result == 2
df = DataFrame(np.random.randn(5, 3))
df2 = DataFrame(np.random.randn(5, 3))
# don't have the pandas parser
with pytest.raises(SyntaxError):
df.query('(@df>0) & (@df2>0)', engine=engine, parser=parser)
with pytest.raises(UndefinedVariableError):
df.query('(df>0) & (df2>0)', engine=engine, parser=parser)
expected = df[(df > 0) & (df2 > 0)]
result = pd.eval('df[(df > 0) & (df2 > 0)]', engine=engine,
parser=parser)
assert_frame_equal(expected, result)
expected = df[(df > 0) & (df2 > 0) & (df[df > 0] > 0)]
result = pd.eval('df[(df > 0) & (df2 > 0) & (df[df > 0] > 0)]',
engine=engine, parser=parser)
assert_frame_equal(expected, result)
class TestDataFrameQueryPythonPandas(TestDataFrameQueryNumExprPandas):
@classmethod
def setup_class(cls):
super(TestDataFrameQueryPythonPandas, cls).setup_class()
cls.engine = 'python'
cls.parser = 'pandas'
cls.frame = TestData().frame
def test_query_builtin(self):
engine, parser = self.engine, self.parser
n = m = 10
df = DataFrame(np.random.randint(m, size=(n, 3)), columns=list('abc'))
df.index.name = 'sin'
expected = df[df.index > 5]
result = df.query('sin > 5', engine=engine, parser=parser)
assert_frame_equal(expected, result)
class TestDataFrameQueryPythonPython(TestDataFrameQueryNumExprPython):
@classmethod
def setup_class(cls):
super(TestDataFrameQueryPythonPython, cls).setup_class()
cls.engine = cls.parser = 'python'
cls.frame = TestData().frame
def test_query_builtin(self):
engine, parser = self.engine, self.parser
n = m = 10
df = DataFrame(np.random.randint(m, size=(n, 3)), columns=list('abc'))
df.index.name = 'sin'
expected = df[df.index > 5]
result = df.query('sin > 5', engine=engine, parser=parser)
assert_frame_equal(expected, result)
class TestDataFrameQueryStrings(object):
def test_str_query_method(self, parser, engine):
tm.skip_if_no_ne(engine)
df = DataFrame(randn(10, 1), columns=['b'])
df['strings'] = Series(list('aabbccddee'))
expect = df[df.strings == 'a']
if parser != 'pandas':
col = 'strings'
lst = '"a"'
lhs = [col] * 2 + [lst] * 2
rhs = lhs[::-1]
eq, ne = '==', '!='
ops = 2 * ([eq] + [ne])
for lhs, op, rhs in zip(lhs, ops, rhs):
ex = '{lhs} {op} {rhs}'.format(lhs=lhs, op=op, rhs=rhs)
pytest.raises(NotImplementedError, df.query, ex,
engine=engine, parser=parser,
local_dict={'strings': df.strings})
else:
res = df.query('"a" == strings', engine=engine, parser=parser)
assert_frame_equal(res, expect)
res = df.query('strings == "a"', engine=engine, parser=parser)
assert_frame_equal(res, expect)
assert_frame_equal(res, df[df.strings.isin(['a'])])
expect = df[df.strings != 'a']
res = df.query('strings != "a"', engine=engine, parser=parser)
assert_frame_equal(res, expect)
res = df.query('"a" != strings', engine=engine, parser=parser)
assert_frame_equal(res, expect)
assert_frame_equal(res, df[~df.strings.isin(['a'])])
def test_str_list_query_method(self, parser, engine):
tm.skip_if_no_ne(engine)
df = DataFrame(randn(10, 1), columns=['b'])
df['strings'] = Series(list('aabbccddee'))
expect = df[df.strings.isin(['a', 'b'])]
if parser != 'pandas':
col = 'strings'
lst = '["a", "b"]'
lhs = [col] * 2 + [lst] * 2
rhs = lhs[::-1]
eq, ne = '==', '!='
ops = 2 * ([eq] + [ne])
for lhs, op, rhs in zip(lhs, ops, rhs):
ex = '{lhs} {op} {rhs}'.format(lhs=lhs, op=op, rhs=rhs)
with pytest.raises(NotImplementedError):
df.query(ex, engine=engine, parser=parser)
else:
res = df.query('strings == ["a", "b"]', engine=engine,
parser=parser)
assert_frame_equal(res, expect)
res = df.query('["a", "b"] == strings', engine=engine,
parser=parser)
assert_frame_equal(res, expect)
expect = df[~df.strings.isin(['a', 'b'])]
res = df.query('strings != ["a", "b"]', engine=engine,
parser=parser)
assert_frame_equal(res, expect)
res = df.query('["a", "b"] != strings', engine=engine,
parser=parser)
assert_frame_equal(res, expect)
def test_query_with_string_columns(self, parser, engine):
tm.skip_if_no_ne(engine)
df = DataFrame({'a': list('aaaabbbbcccc'),
'b': list('aabbccddeeff'),
'c': np.random.randint(5, size=12),
'd': np.random.randint(9, size=12)})
if parser == 'pandas':
res = df.query('a in b', parser=parser, engine=engine)
expec = df[df.a.isin(df.b)]
assert_frame_equal(res, expec)
res = df.query('a in b and c < d', parser=parser, engine=engine)
expec = df[df.a.isin(df.b) & (df.c < df.d)]
assert_frame_equal(res, expec)
else:
with pytest.raises(NotImplementedError):
df.query('a in b', parser=parser, engine=engine)
with pytest.raises(NotImplementedError):
df.query('a in b and c < d', parser=parser, engine=engine)
def test_object_array_eq_ne(self, parser, engine):
tm.skip_if_no_ne(engine)
df = DataFrame({'a': list('aaaabbbbcccc'),
'b': list('aabbccddeeff'),
'c': np.random.randint(5, size=12),
'd': np.random.randint(9, size=12)})
res = df.query('a == b', parser=parser, engine=engine)
exp = df[df.a == df.b]
assert_frame_equal(res, exp)
res = df.query('a != b', parser=parser, engine=engine)
exp = df[df.a != df.b]
assert_frame_equal(res, exp)
def test_query_with_nested_strings(self, parser, engine):
tm.skip_if_no_ne(engine)
skip_if_no_pandas_parser(parser)
raw = """id event timestamp
1 "page 1 load" 1/1/2014 0:00:01
1 "page 1 exit" 1/1/2014 0:00:31
2 "page 2 load" 1/1/2014 0:01:01
2 "page 2 exit" 1/1/2014 0:01:31
3 "page 3 load" 1/1/2014 0:02:01
3 "page 3 exit" 1/1/2014 0:02:31
4 "page 1 load" 2/1/2014 1:00:01
4 "page 1 exit" 2/1/2014 1:00:31
5 "page 2 load" 2/1/2014 1:01:01
5 "page 2 exit" 2/1/2014 1:01:31
6 "page 3 load" 2/1/2014 1:02:01
6 "page 3 exit" 2/1/2014 1:02:31
"""
df = pd.read_csv(StringIO(raw), sep=r'\s{2,}', engine='python',
parse_dates=['timestamp'])
expected = df[df.event == '"page 1 load"']
res = df.query("""'"page 1 load"' in event""", parser=parser,
engine=engine)
assert_frame_equal(expected, res)
def test_query_with_nested_special_character(self, parser, engine):
skip_if_no_pandas_parser(parser)
tm.skip_if_no_ne(engine)
df = DataFrame({'a': ['a', 'b', 'test & test'],
'b': [1, 2, 3]})
res = df.query('a == "test & test"', parser=parser, engine=engine)
expec = df[df.a == 'test & test']
assert_frame_equal(res, expec)
def test_query_lex_compare_strings(self, parser, engine):
tm.skip_if_no_ne(engine=engine)
import operator as opr
a = Series(np.random.choice(list('abcde'), 20))
b = Series(np.arange(a.size))
df = DataFrame({'X': a, 'Y': b})
ops = {'<': opr.lt, '>': opr.gt, '<=': opr.le, '>=': opr.ge}
for op, func in ops.items():
res = df.query('X %s "d"' % op, engine=engine, parser=parser)
expected = df[func(df.X, 'd')]
assert_frame_equal(res, expected)
def test_query_single_element_booleans(self, parser, engine):
tm.skip_if_no_ne(engine)
columns = 'bid', 'bidsize', 'ask', 'asksize'
data = np.random.randint(2, size=(1, len(columns))).astype(bool)
df = DataFrame(data, columns=columns)
res = df.query('bid & ask', engine=engine, parser=parser)
expected = df[df.bid & df.ask]
assert_frame_equal(res, expected)
def test_query_string_scalar_variable(self, parser, engine):
tm.skip_if_no_ne(engine)
skip_if_no_pandas_parser(parser)
df = pd.DataFrame({'Symbol': ['BUD US', 'BUD US', 'IBM US', 'IBM US'],
'Price': [109.70, 109.72, 183.30, 183.35]})
e = df[df.Symbol == 'BUD US']
symb = 'BUD US' # noqa
r = df.query('Symbol == @symb', parser=parser, engine=engine)
assert_frame_equal(e, r)
class TestDataFrameEvalNumExprPandas(object):
@classmethod
def setup_class(cls):
cls.engine = 'numexpr'
cls.parser = 'pandas'
tm.skip_if_no_ne()
def setup_method(self, method):
self.frame = DataFrame(randn(10, 3), columns=list('abc'))
def teardown_method(self, method):
del self.frame
def test_simple_expr(self):
res = self.frame.eval('a + b', engine=self.engine, parser=self.parser)
expect = self.frame.a + self.frame.b
assert_series_equal(res, expect)
def test_bool_arith_expr(self):
res = self.frame.eval('a[a < 1] + b', engine=self.engine,
parser=self.parser)
expect = self.frame.a[self.frame.a < 1] + self.frame.b
assert_series_equal(res, expect)
def test_invalid_type_for_operator_raises(self):
df = DataFrame({'a': [1, 2], 'b': ['c', 'd']})
ops = '+', '-', '*', '/'
for op in ops:
with tm.assert_raises_regex(TypeError,
"unsupported operand type\(s\) "
"for .+: '.+' and '.+'"):
df.eval('a {0} b'.format(op), engine=self.engine,
parser=self.parser)
class TestDataFrameEvalNumExprPython(TestDataFrameEvalNumExprPandas):
@classmethod
def setup_class(cls):
super(TestDataFrameEvalNumExprPython, cls).setup_class()
cls.engine = 'numexpr'
cls.parser = 'python'
tm.skip_if_no_ne(cls.engine)
class TestDataFrameEvalPythonPandas(TestDataFrameEvalNumExprPandas):
@classmethod
def setup_class(cls):
super(TestDataFrameEvalPythonPandas, cls).setup_class()
cls.engine = 'python'
cls.parser = 'pandas'
class TestDataFrameEvalPythonPython(TestDataFrameEvalNumExprPython):
@classmethod
def setup_class(cls):
cls.engine = cls.parser = 'python'
| mit |
jniediek/mne-python | examples/visualization/plot_evoked_topomap.py | 13 | 1606 | """
========================================
Plotting topographic maps of evoked data
========================================
Load evoked data and plot topomaps for selected time points.
"""
# Authors: Christian Brodbeck <christianbrodbeck@nyu.edu>
# Tal Linzen <linzen@nyu.edu>
# Denis A. Engeman <denis.engemann@gmail.com>
#
# License: BSD (3-clause)
import numpy as np
import matplotlib.pyplot as plt
from mne.datasets import sample
from mne import read_evokeds
print(__doc__)
path = sample.data_path()
fname = path + '/MEG/sample/sample_audvis-ave.fif'
# load evoked and subtract baseline
condition = 'Left Auditory'
evoked = read_evokeds(fname, condition=condition, baseline=(None, 0))
# set time instants in seconds (from 50 to 150ms in a step of 10ms)
times = np.arange(0.05, 0.15, 0.01)
# If times is set to None only 10 regularly spaced topographies will be shown
# plot magnetometer data as topomaps
evoked.plot_topomap(times, ch_type='mag')
# compute a 50 ms bin to stabilize topographies
evoked.plot_topomap(times, ch_type='mag', average=0.05)
# plot gradiometer data (plots the RMS for each pair of gradiometers)
evoked.plot_topomap(times, ch_type='grad')
# plot magnetometer data as an animation
evoked.animate_topomap(ch_type='mag', times=times, frame_rate=10)
# plot magnetometer data as topomap at 1 time point : 100 ms
# and add channel labels and title
evoked.plot_topomap(0.1, ch_type='mag', show_names=True, colorbar=False,
size=6, res=128, title='Auditory response')
plt.subplots_adjust(left=0.01, right=0.99, bottom=0.01, top=0.88)
| bsd-3-clause |
RPGOne/Skynet | 5230d93ccc9fa5329b0a02a351b02939-459eebff35e625675d2f6ff5633c7051c1d64a0e/gistfile1.py | 1 | 3974 | """
python speedup_kmeans.py --profile
python speedup_kmeans.py
git worktree add workdir_master master
rob sedr "\<sklearn\>" sklearn_master True
git mv sklearn sklearn_master
python setup develop
python -c "import sklearn_master; print(sklearn_master.__file__)"
python -c "import sklearn; print(sklearn.__file__)"
"""
from __future__ import absolute_import, division, print_function, unicode_literals
import utool as ut
import sklearn # NOQA
from sklearn.datasets.samples_generator import make_blobs
from sklearn.utils.extmath import row_norms, squared_norm # NOQA
import sklearn.cluster
import numpy as np
from sklearn.metrics.pairwise import euclidean_distances # NOQA
import sklearn_master.cluster
(print, rrr, profile) = ut.inject2(__name__, '[tester]')
def test_kmeans_plus_plus_speed(n_clusters=2000, n_features=128, per_cluster=10, asint=False, fix=True):
"""
from speedup_kmeans import *
from sklearn.cluster.k_means_ import *
"""
rng = np.random.RandomState(42)
# Make random cluster centers on a ball
centers = rng.rand(n_clusters, n_features)
centers /= np.linalg.norm(centers, axis=0)[None, :]
centers = (centers * 512).astype(np.uint8) / 512
centers /= np.linalg.norm(centers, axis=0)[None, :]
n_samples = int(n_clusters * per_cluster)
n_clusters, n_features = centers.shape
X, true_labels = make_blobs(n_samples=n_samples, centers=centers,
cluster_std=1., random_state=42)
if asint:
X = (X * 512).astype(np.int32)
x_squared_norms = row_norms(X, squared=True)
if fix:
_k_init = sklearn.cluster.k_means_._k_init
else:
_k_init = sklearn_master.cluster.k_means_._k_init
random_state = np.random.RandomState(42)
n_local_trials = None # NOQA
with ut.Timer('testing kmeans init') as t:
centers = _k_init(X, n_clusters, random_state=random_state, x_squared_norms=x_squared_norms)
return centers, t.ellapsed
def main():
if True:
import pandas as pd
pd.options.display.max_rows = 1000
pd.options.display.width = 1000
basis = {
#'n_clusters': [10, 100, 1000, 2000][::-1],
#'n_features': [4, 32, 128, 512][::-1],
#'per_cluster': [1, 10, 100, 200][::-1],
'n_clusters': [10, 100, 500][::-1],
'n_features': [32, 128][::-1],
'per_cluster': [1, 10, 20][::-1],
'asint': [True, False],
}
vals = []
for kw in ut.ProgIter(ut.all_dict_combinations(basis), lbl='gridsearch',
bs=False, adjust=False, freq=1):
print('kw = ' + ut.repr2(kw))
exec(ut.execstr_dict(kw))
centers1, new_speed = test_kmeans_plus_plus_speed(fix=True, **kw)
centers2, old_speed = test_kmeans_plus_plus_speed(fix=False, **kw)
import utool
with utool.embed_on_exception_context:
assert np.all(centers1 == centers2), 'new code disagrees'
kw['new_speed'] = new_speed
kw['old_speed'] = old_speed
vals.append(kw)
print('---------')
df = pd.DataFrame.from_dict(vals)
df['percent_change'] = 100 * (df['old_speed'] - df['new_speed']) / df['old_speed']
df = df.reindex_axis(list(basis.keys()) + ['new_speed', 'old_speed', 'percent_change'], axis=1)
df['absolute_change'] = (df['old_speed'] - df['new_speed'])
print(df.sort('absolute_change', ascending=False))
#print(df)
print(df['percent_change'][df['absolute_change'] > .1].mean())
#print(df.loc[df['percent_change'].argsort()[::-1]])
else:
new_speed = test_kmeans_plus_plus_speed()
try:
profile.dump_stats('out.lprof')
profile.print_stats(stripzeros=True)
except Exception:
pass
print('new_speed = %r' % (new_speed,))
if __name__ == '__main__':
main()
| bsd-3-clause |
JoeBartelmo/PyDetect | gui/img_proc/GlobalSurveyor.py | 2 | 10667 | # Copyright (c) 2016, Jeffrey Maggio and Joseph Bartelmo
#
# Permission is hereby granted, free of charge, to any person obtaining a copy of this software and
# associated documentation files (the "Software"), to deal in the Software without restriction,
# including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense,
# and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so,
# subject to the following conditions:
#
# The above copyright notice and this permission notice shall be included in all copies or substantial
# portions of the Software.
#
# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT
# LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT.
# IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
# LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION
# WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
import Tkinter as tk
from PIL import Image, ImageTk
import cv2
import sys
import numpy
import time
import matplotlib.path as matpath
class GlobalSurveyor(object):
def __init__(self, root, ideal_image, number_polys = 2, thresholds = (50, 200), polys = None):
self.ideal_im = ideal_image
self.parent = root
self.done = False
self.points = []
self.warningColor = (128, 128, 0)
self.errorColor = (255, 0, 0)
self.number_polys = number_polys
#self.number_sides = number_sides
if polys == None:
self.polys = []
else:
self.polys = polys
self.thresh = thresholds
self.ideal_counts = []
self.calibrates_polygons()
def calibrates_polygons(self):
polys = self.define_polygons(self.ideal_im)
ideal_kp = self.apply_fast(self.ideal_im)
self.ideal_counts = self.keypoints_to_counts(ideal_kp)
def _on_left_mouse(self, event): # CITE THIS FROM STACKEXCHANGE
# Mouse callback that gets called for every mouse event (i.e. moving, clicking, etc.)
if self.done: # Nothing more to do
return
# Left click means adding a point at current position to the list of points
print("Adding point #%d with position(%d,%d)" % (len(self.points), event.x, event.y))
self.points.append((event.x, event.y))
def _on_right_mouse(self, event): # CITE THIS FROM STACKEXCHANGE
if len(self.points) > 2:
# Right click means we're done
print("Completing polygon with %d points." % len(self.points))
self.done = True
def define_polygons(self, image):
# use this to automate point picking and/or display point picker window in gui
FINAL_LINE_COLOR = (255, 0, 0)
WORKING_LINE_COLOR = (0, 0, 255)
top = tk.Toplevel(self.parent)
top.title("Select bounding points for the target area")
print image.shape
top.update_idletasks()
width = image.shape[1]
height = image.shape[0]
x = (top.winfo_screenwidth() // 2) - (width // 2)
y = (top.winfo_screenheight() // 2) - (height // 2)
top.geometry('{}x{}+{}+{}'.format(width, height, x, y))
#we are defaulting to the image size, i don't want to deal wiht resizing
initial_im = tk.PhotoImage()
image_label = tk.Label(top, image=initial_im)
image_label.grid(row = 0, column = 0)
image_label.bind("<Button-1>", self._on_left_mouse)
image_label.bind("<Button-3>", self._on_right_mouse)
top.grid()
copy = image.copy()
def display_image():
imageFromArray = Image.fromarray(copy)
try:
tkImage = ImageTk.PhotoImage(image=imageFromArray)
image_label.configure(image=tkImage)
image_label._image_cache = tkImage # avoid garbage collection
top.update()
return True
except RuntimeError:
print('Unable to update image frame. Assuming application has been killed unexpectidly.')
return False
for num in range(self.number_polys):
if display_image() == False:
top.destroy()
return
self.points = []
self.done = False
while (not self.done):
if (len(self.points) > 0):
cv2.polylines(copy, numpy.asarray([self.points]), False, FINAL_LINE_COLOR, 1)
imageFromArray = Image.fromarray(copy)
if display_image() == False:
top.destroy()
return
self.polys.append(self.points)
print(self.polys)
update = numpy.asarray(self.polys[num]).astype(numpy.int32)
if (len(self.points) > 0):
cv2.fillPoly(copy, [update], (0,0,255))
display_image()
time.sleep(0.5)
top.destroy()
return self.polys
def apply_fast(self, image):
fast = cv2.FastFeatureDetector()
kp = fast.detect(image, None)
return kp
def keypoints_to_counts(self, im_keypoints): #polys = numpy.asarray[[[0,0],[0,ideal_im.shape[0]//2],[ideal_im.shape[0]//2,ideal_im.shape[1]],[ideal_im.shape[0]//2, 0]]]):
#pass in im_keypoints and count how many are in each polygon
#polygon points should also be an input
im_keys = []
for i in range(len(im_keypoints)):
im_keys.append(im_keypoints[i].pt)
# NOTE:: POLYS MUST BE IN X (COL), Y (ROW) FORMAT FOR FILLPOLY TO WORK
# NOTE:: POLYS MUST ALSO BE OF DTYPE FLOAT
data = self.polys
polys = map(lambda data: map(lambda data: map(float, data), data), data)
counts = []
for poly in polys:
# MATPLOTLIB PATH EXAMPLE GOES HERE
bbPath = matpath.Path(poly)
# sum boolean array to get number of kp within current poly
try:
truth_vector = bbPath.contains_points(im_keys) # check input for contains_points. might only like tuples
except:
truth_vector = [1] * len(im_keys)
counts.append(numpy.sum(truth_vector))
return numpy.asarray(counts)
def threshold(self, counts, thresh_tuple):
# do the thresholding thing
delta = abs(counts - self.ideal_counts)
threshed_polys = numpy.where((delta >= thresh_tuple[0]) & (delta < thresh_tuple[1]))[0] # ok to index like this because tuple represents vector
# save which polys get marked true out so these can be used as overlay in fillpolys
# this can be done by indexing into polys with truth vector saved here
return threshed_polys.astype(int) # a true/false vector of length polys
def generate_output(self, image, threshed_polys, color, alpha = 0.5): #, colorAlert = (0,0,255)):
# use polyfill to blend im_in and overlay (which is just the polygons we want colored in)
colored_polys = []
for threshed_poly in threshed_polys:
colored_polys.append(numpy.array(self.polys[threshed_poly]).astype(numpy.int32))
overlay = image.copy()
im_out = image.copy()
for cp in range(len(colored_polys)):
cv2.fillPoly(overlay, [colored_polys[cp]], color)
cv2.addWeighted(overlay, alpha, im_out, 1 - alpha, 0, im_out)
return im_out
def run_basic_fod(self, current_im):
if current_im is None:
return None
# load ideal image
im_in_kp = self.apply_fast(current_im)
counts = self.keypoints_to_counts(im_keypoints = im_in_kp)
warning_polys = self.threshold(counts, self.thresh)
error_polys = self.threshold(counts, (self.thresh[1], sys.maxint))
warning_overlay = self.generate_output(current_im, warning_polys, self.warningColor)
return self.generate_output(warning_overlay, error_polys, self.errorColor)
def get_thresholds_widget(parent, values_list):
top = tk.Toplevel(parent)
top.title("Select your desired thresholds")
top.update_idletasks()
thresh1 = tk.IntVar()
thresh2 = tk.IntVar()
polys = tk.IntVar()
#3 labels and 3 textbox selectors
label1 = tk.Label(top, text = "Warning (Maybe an object)")
label1.grid(row = 1, column = 0, sticky ="nsew")
threshold1 = tk.Entry(top, textvariable=thresh1)
threshold1.grid(row = 1, column = 1, sticky ="nsew")
label2 = tk.Label(top, text = "Error (Most defintely an object)")
label2.grid(row = 2, column = 0, sticky ="nsew")
threshold2 = tk.Entry(top, textvariable = thresh2)
threshold2.grid(row = 2, column = 1, sticky ="nsew")
label3 = tk.Label(top, text = "Number of Polygons")
label3.grid(row = 3, column = 0, sticky ="nsew")
threshold3 = tk.Entry(top, textvariable = polys)
threshold3.grid(row = 3, column = 1, sticky ="nsew")
def returnVars():
top.destroy()
if len(values_list) != 3:
values_list.append(thresh1.get())
values_list.append(thresh2.get())
values_list.append(polys.get())
else:
values_list[0] = thresh1.get()
values_list[1] = thresh2.get()
values_list[2] = polys.get()
button = tk.Button(top, text = "Set Values", command = returnVars)
button.grid(row = 4, column = 0, columnspan = 2, sticky ="nsew")
threshold1.delete(0, "end")
threshold2.delete(0, "end")
threshold3.delete(0, "end")
if len(values_list) == 3:
threshold1.insert(0, values_list[0])
threshold2.insert(0, values_list[1])
threshold3.insert(0, values_list[2])
else:
threshold1.insert(0, 50)
threshold2.insert(0, 200)
threshold3.insert(0, 2)
width = 300
height = 100
x = (top.winfo_screenwidth() // 2) - (width // 2)
y = (top.winfo_screenheight() // 2) - (height // 2)
top.geometry('{}x{}+{}+{}'.format(width, height, x, y))
top.grid_columnconfigure(1, weight=1)
top.grid_rowconfigure(0, weight=1)
top.grid_rowconfigure(1, weight=1)
top.grid()
top.update()
parent.wait_window(top)
if __name__=='__main__':
root = tk.Tk()
vals = []
get_thresholds_widget(root, vals)
print 'obtained values', vals
get_thresholds_widget(root, vals)
print 'obtained values', vals
root.mainloop()
| mit |
michigraber/scikit-learn | sklearn/decomposition/truncated_svd.py | 199 | 7744 | """Truncated SVD for sparse matrices, aka latent semantic analysis (LSA).
"""
# Author: Lars Buitinck <L.J.Buitinck@uva.nl>
# Olivier Grisel <olivier.grisel@ensta.org>
# Michael Becker <mike@beckerfuffle.com>
# License: 3-clause BSD.
import numpy as np
import scipy.sparse as sp
try:
from scipy.sparse.linalg import svds
except ImportError:
from ..utils.arpack import svds
from ..base import BaseEstimator, TransformerMixin
from ..utils import check_array, as_float_array, check_random_state
from ..utils.extmath import randomized_svd, safe_sparse_dot, svd_flip
from ..utils.sparsefuncs import mean_variance_axis
__all__ = ["TruncatedSVD"]
class TruncatedSVD(BaseEstimator, TransformerMixin):
"""Dimensionality reduction using truncated SVD (aka LSA).
This transformer performs linear dimensionality reduction by means of
truncated singular value decomposition (SVD). It is very similar to PCA,
but operates on sample vectors directly, instead of on a covariance matrix.
This means it can work with scipy.sparse matrices efficiently.
In particular, truncated SVD works on term count/tf-idf matrices as
returned by the vectorizers in sklearn.feature_extraction.text. In that
context, it is known as latent semantic analysis (LSA).
This estimator supports two algorithm: a fast randomized SVD solver, and
a "naive" algorithm that uses ARPACK as an eigensolver on (X * X.T) or
(X.T * X), whichever is more efficient.
Read more in the :ref:`User Guide <LSA>`.
Parameters
----------
n_components : int, default = 2
Desired dimensionality of output data.
Must be strictly less than the number of features.
The default value is useful for visualisation. For LSA, a value of
100 is recommended.
algorithm : string, default = "randomized"
SVD solver to use. Either "arpack" for the ARPACK wrapper in SciPy
(scipy.sparse.linalg.svds), or "randomized" for the randomized
algorithm due to Halko (2009).
n_iter : int, optional
Number of iterations for randomized SVD solver. Not used by ARPACK.
random_state : int or RandomState, optional
(Seed for) pseudo-random number generator. If not given, the
numpy.random singleton is used.
tol : float, optional
Tolerance for ARPACK. 0 means machine precision. Ignored by randomized
SVD solver.
Attributes
----------
components_ : array, shape (n_components, n_features)
explained_variance_ratio_ : array, [n_components]
Percentage of variance explained by each of the selected components.
explained_variance_ : array, [n_components]
The variance of the training samples transformed by a projection to
each component.
Examples
--------
>>> from sklearn.decomposition import TruncatedSVD
>>> from sklearn.random_projection import sparse_random_matrix
>>> X = sparse_random_matrix(100, 100, density=0.01, random_state=42)
>>> svd = TruncatedSVD(n_components=5, random_state=42)
>>> svd.fit(X) # doctest: +NORMALIZE_WHITESPACE
TruncatedSVD(algorithm='randomized', n_components=5, n_iter=5,
random_state=42, tol=0.0)
>>> print(svd.explained_variance_ratio_) # doctest: +ELLIPSIS
[ 0.07825... 0.05528... 0.05445... 0.04997... 0.04134...]
>>> print(svd.explained_variance_ratio_.sum()) # doctest: +ELLIPSIS
0.27930...
See also
--------
PCA
RandomizedPCA
References
----------
Finding structure with randomness: Stochastic algorithms for constructing
approximate matrix decompositions
Halko, et al., 2009 (arXiv:909) http://arxiv.org/pdf/0909.4061
Notes
-----
SVD suffers from a problem called "sign indeterminancy", which means the
sign of the ``components_`` and the output from transform depend on the
algorithm and random state. To work around this, fit instances of this
class to data once, then keep the instance around to do transformations.
"""
def __init__(self, n_components=2, algorithm="randomized", n_iter=5,
random_state=None, tol=0.):
self.algorithm = algorithm
self.n_components = n_components
self.n_iter = n_iter
self.random_state = random_state
self.tol = tol
def fit(self, X, y=None):
"""Fit LSI model on training data X.
Parameters
----------
X : {array-like, sparse matrix}, shape (n_samples, n_features)
Training data.
Returns
-------
self : object
Returns the transformer object.
"""
self.fit_transform(X)
return self
def fit_transform(self, X, y=None):
"""Fit LSI model to X and perform dimensionality reduction on X.
Parameters
----------
X : {array-like, sparse matrix}, shape (n_samples, n_features)
Training data.
Returns
-------
X_new : array, shape (n_samples, n_components)
Reduced version of X. This will always be a dense array.
"""
X = as_float_array(X, copy=False)
random_state = check_random_state(self.random_state)
# If sparse and not csr or csc, convert to csr
if sp.issparse(X) and X.getformat() not in ["csr", "csc"]:
X = X.tocsr()
if self.algorithm == "arpack":
U, Sigma, VT = svds(X, k=self.n_components, tol=self.tol)
# svds doesn't abide by scipy.linalg.svd/randomized_svd
# conventions, so reverse its outputs.
Sigma = Sigma[::-1]
U, VT = svd_flip(U[:, ::-1], VT[::-1])
elif self.algorithm == "randomized":
k = self.n_components
n_features = X.shape[1]
if k >= n_features:
raise ValueError("n_components must be < n_features;"
" got %d >= %d" % (k, n_features))
U, Sigma, VT = randomized_svd(X, self.n_components,
n_iter=self.n_iter,
random_state=random_state)
else:
raise ValueError("unknown algorithm %r" % self.algorithm)
self.components_ = VT
# Calculate explained variance & explained variance ratio
X_transformed = np.dot(U, np.diag(Sigma))
self.explained_variance_ = exp_var = np.var(X_transformed, axis=0)
if sp.issparse(X):
_, full_var = mean_variance_axis(X, axis=0)
full_var = full_var.sum()
else:
full_var = np.var(X, axis=0).sum()
self.explained_variance_ratio_ = exp_var / full_var
return X_transformed
def transform(self, X):
"""Perform dimensionality reduction on X.
Parameters
----------
X : {array-like, sparse matrix}, shape (n_samples, n_features)
New data.
Returns
-------
X_new : array, shape (n_samples, n_components)
Reduced version of X. This will always be a dense array.
"""
X = check_array(X, accept_sparse='csr')
return safe_sparse_dot(X, self.components_.T)
def inverse_transform(self, X):
"""Transform X back to its original space.
Returns an array X_original whose transform would be X.
Parameters
----------
X : array-like, shape (n_samples, n_components)
New data.
Returns
-------
X_original : array, shape (n_samples, n_features)
Note that this is always a dense array.
"""
X = check_array(X)
return np.dot(X, self.components_)
| bsd-3-clause |
wataash/Instr | instr/ke2636a.py | 1 | 3794 | import numpy as np
import unittest2
from instr.base import SourceMeter
class Keithley2636A(SourceMeter):
def __init__(self, rsrc=None, timeout_sec=600, reset=True):
self._smu = 'a'
idn = 'Keithley Instruments Inc., Model 2636A'
super().__init__(rsrc, idn, timeout_sec, reset)
@property
def smu(self):
return self._smu
@smu.setter
def smu(self, value):
if value not in ['a', 'b']:
raise ValueError
self._smu = value
def check_error(self):
if self._debug_mode:
super().check_error()
tmp = self.q('print(errorqueue.next())')
if tmp != '0.00000e+00\tQueue Is Empty\t0.00000e+00\n':
raise RuntimeError('Error on Keithley 2636A.')
def reset(self):
self.w('reset()', True)
# self.w('smua.reset(); smub.reset()', True)
def iv_sweep(self, v_start=0.0, v_end=10e-3, v_step=1e-3,
v_points=None, i_limit=1e-6, settle_time=0.0, reset=True):
"""
Reference manual 3-31
TODO: when aborted?
:return: vis, is_aborted
"""
if reset:
self.reset()
if v_points is None:
v_points = self._v_step_to_points(v_start, v_end, v_step)
lim = 'smu{}.source.limiti = {}'.format(self.smu, i_limit)
self.w(lim, True)
meas = 'SweepVLinMeasureI(smu{}, {}, {}, {}, {})'. \
format(self.smu, v_start, v_end, settle_time, v_points)
self.w(meas, True)
prnt = 'printbuffer(1, {}, smu{}.nvbuffer1.readings)'. \
format(v_points, self.smu)
resp = self.q(prnt, True)
Is = resp.split(', ')
Is = np.asarray(Is, np.float64)
if len(Is) != v_points:
aborted = True
v_points = len(Is)
else:
aborted = False
vs = np.linspace(v_start, v_end, v_points)
vis = np.array([vs, Is]).transpose()
return vis, aborted
def iv_sweep_double(self, v_max, v_step=1e-3, v_points=None,
i_limit=1e-3, settle_time=0.0, reset=True):
vis1, aborted = self.iv_sweep(0, v_max, v_step,
v_points, i_limit, settle_time, reset)
if aborted:
return vis1, aborted
vis2, aborted = self.iv_sweep(v_max, 0, v_step,
v_points, i_limit, settle_time, reset)
ret = np.concatenate((vis1, vis2))
return ret, aborted
class TestKeithley2636A(unittest2.TestCase):
def test_iv_sweep(self):
import matplotlib.pyplot as plt
ke2636a.reset()
v_start = 0.0
v_end = 1e-3
self.smu = 'a'
vis, aborted = \
ke2636a.iv_sweep(v_start, v_end, v_step=v_end / 10, i_limit=1e-9)
plt.plot(*vis.transpose(), 'o-')
plt.show()
vis, aborted = \
ke2636a.iv_sweep(v_start, v_end, v_points=101, i_limit=1e-6)
plt.plot(*vis.transpose(), 'o-')
plt.show()
# v_step ignored
vis, aborted = \
ke2636a.iv_sweep(v_start, v_end, v_step=1, v_points=11)
plt.plot(*vis.transpose(), 'o-')
plt.show()
vis, aborted = ke2636a.iv_sweep_double(10e-3)
plt.plot(*vis.transpose(), 'o-')
plt.show()
self.smu = 'b'
vis, aborted = ke2636a.iv_sweep(v_start, v_end, v_points=11)
plt.plot(*vis.transpose(), 'o-')
plt.show()
if __name__ == '__main__':
import visa
rm = visa.ResourceManager()
# ke2636a_rsrc = rm.open_resource('visa://169.254.136.196/GPIB0::20::INSTR')
ke2636a_rsrc = rm.open_resource('TCPIP::169.254.000.001::INSTR')
ke2636a = Keithley2636A(ke2636a_rsrc)
unittest2.main()
pass
| mit |
arank/mxnet | example/reinforcement-learning/ddpg/strategies.py | 15 | 1705 | import numpy as np
class BaseStrategy(object):
"""
Base class of exploration strategy.
"""
def get_action(self, obs, policy):
raise NotImplementedError
def reset(self):
pass
class OUStrategy(BaseStrategy):
"""
Ornstein-Uhlenbeck process: dxt = theta * (mu - xt) * dt + sigma * dWt
where Wt denotes the Wiener process.
"""
def __init__(self, env_spec, mu=0, theta=0.15, sigma=0.3):
self.mu = mu
self.theta = theta
self.sigma = sigma
self.action_space = env_spec.action_space
self.state = np.ones(self.action_space.flat_dim) * self.mu
def evolve_state(self):
x = self.state
dx = self.theta * (self.mu - x) + self.sigma * np.random.randn(len(x))
self.state = x + dx
return self.state
def reset(self):
self.state = np.ones(self.action_space.flat_dim) * self.mu
def get_action(self, obs, policy):
# get_action accepts a 2D tensor with one row
obs = obs.reshape((1, -1))
action = policy.get_action(obs)
increment = self.evolve_state()
return np.clip(action + increment,
self.action_space.low,
self.action_space.high)
if __name__ == "__main__":
class Env1(object):
def __init__(self):
self.action_space = Env2()
class Env2(object):
def __init__(self):
self.flat_dim = 2
env_spec = Env1()
test = OUStrategy(env_spec)
states = []
for i in range(1000):
states.append(test.evolve_state()[0])
import matplotlib.pyplot as plt
plt.plot(states)
plt.show()
| apache-2.0 |
frank-tancf/scikit-learn | benchmarks/bench_multilabel_metrics.py | 276 | 7138 | #!/usr/bin/env python
"""
A comparison of multilabel target formats and metrics over them
"""
from __future__ import division
from __future__ import print_function
from timeit import timeit
from functools import partial
import itertools
import argparse
import sys
import matplotlib.pyplot as plt
import scipy.sparse as sp
import numpy as np
from sklearn.datasets import make_multilabel_classification
from sklearn.metrics import (f1_score, accuracy_score, hamming_loss,
jaccard_similarity_score)
from sklearn.utils.testing import ignore_warnings
METRICS = {
'f1': partial(f1_score, average='micro'),
'f1-by-sample': partial(f1_score, average='samples'),
'accuracy': accuracy_score,
'hamming': hamming_loss,
'jaccard': jaccard_similarity_score,
}
FORMATS = {
'sequences': lambda y: [list(np.flatnonzero(s)) for s in y],
'dense': lambda y: y,
'csr': lambda y: sp.csr_matrix(y),
'csc': lambda y: sp.csc_matrix(y),
}
@ignore_warnings
def benchmark(metrics=tuple(v for k, v in sorted(METRICS.items())),
formats=tuple(v for k, v in sorted(FORMATS.items())),
samples=1000, classes=4, density=.2,
n_times=5):
"""Times metric calculations for a number of inputs
Parameters
----------
metrics : array-like of callables (1d or 0d)
The metric functions to time.
formats : array-like of callables (1d or 0d)
These may transform a dense indicator matrix into multilabel
representation.
samples : array-like of ints (1d or 0d)
The number of samples to generate as input.
classes : array-like of ints (1d or 0d)
The number of classes in the input.
density : array-like of ints (1d or 0d)
The density of positive labels in the input.
n_times : int
Time calling the metric n_times times.
Returns
-------
array of floats shaped like (metrics, formats, samples, classes, density)
Time in seconds.
"""
metrics = np.atleast_1d(metrics)
samples = np.atleast_1d(samples)
classes = np.atleast_1d(classes)
density = np.atleast_1d(density)
formats = np.atleast_1d(formats)
out = np.zeros((len(metrics), len(formats), len(samples), len(classes),
len(density)), dtype=float)
it = itertools.product(samples, classes, density)
for i, (s, c, d) in enumerate(it):
_, y_true = make_multilabel_classification(n_samples=s, n_features=1,
n_classes=c, n_labels=d * c,
random_state=42)
_, y_pred = make_multilabel_classification(n_samples=s, n_features=1,
n_classes=c, n_labels=d * c,
random_state=84)
for j, f in enumerate(formats):
f_true = f(y_true)
f_pred = f(y_pred)
for k, metric in enumerate(metrics):
t = timeit(partial(metric, f_true, f_pred), number=n_times)
out[k, j].flat[i] = t
return out
def _tabulate(results, metrics, formats):
"""Prints results by metric and format
Uses the last ([-1]) value of other fields
"""
column_width = max(max(len(k) for k in formats) + 1, 8)
first_width = max(len(k) for k in metrics)
head_fmt = ('{:<{fw}s}' + '{:>{cw}s}' * len(formats))
row_fmt = ('{:<{fw}s}' + '{:>{cw}.3f}' * len(formats))
print(head_fmt.format('Metric', *formats,
cw=column_width, fw=first_width))
for metric, row in zip(metrics, results[:, :, -1, -1, -1]):
print(row_fmt.format(metric, *row,
cw=column_width, fw=first_width))
def _plot(results, metrics, formats, title, x_ticks, x_label,
format_markers=('x', '|', 'o', '+'),
metric_colors=('c', 'm', 'y', 'k', 'g', 'r', 'b')):
"""
Plot the results by metric, format and some other variable given by
x_label
"""
fig = plt.figure('scikit-learn multilabel metrics benchmarks')
plt.title(title)
ax = fig.add_subplot(111)
for i, metric in enumerate(metrics):
for j, format in enumerate(formats):
ax.plot(x_ticks, results[i, j].flat,
label='{}, {}'.format(metric, format),
marker=format_markers[j],
color=metric_colors[i % len(metric_colors)])
ax.set_xlabel(x_label)
ax.set_ylabel('Time (s)')
ax.legend()
plt.show()
if __name__ == "__main__":
ap = argparse.ArgumentParser()
ap.add_argument('metrics', nargs='*', default=sorted(METRICS),
help='Specifies metrics to benchmark, defaults to all. '
'Choices are: {}'.format(sorted(METRICS)))
ap.add_argument('--formats', nargs='+', choices=sorted(FORMATS),
help='Specifies multilabel formats to benchmark '
'(defaults to all).')
ap.add_argument('--samples', type=int, default=1000,
help='The number of samples to generate')
ap.add_argument('--classes', type=int, default=10,
help='The number of classes')
ap.add_argument('--density', type=float, default=.2,
help='The average density of labels per sample')
ap.add_argument('--plot', choices=['classes', 'density', 'samples'],
default=None,
help='Plot time with respect to this parameter varying '
'up to the specified value')
ap.add_argument('--n-steps', default=10, type=int,
help='Plot this many points for each metric')
ap.add_argument('--n-times',
default=5, type=int,
help="Time performance over n_times trials")
args = ap.parse_args()
if args.plot is not None:
max_val = getattr(args, args.plot)
if args.plot in ('classes', 'samples'):
min_val = 2
else:
min_val = 0
steps = np.linspace(min_val, max_val, num=args.n_steps + 1)[1:]
if args.plot in ('classes', 'samples'):
steps = np.unique(np.round(steps).astype(int))
setattr(args, args.plot, steps)
if args.metrics is None:
args.metrics = sorted(METRICS)
if args.formats is None:
args.formats = sorted(FORMATS)
results = benchmark([METRICS[k] for k in args.metrics],
[FORMATS[k] for k in args.formats],
args.samples, args.classes, args.density,
args.n_times)
_tabulate(results, args.metrics, args.formats)
if args.plot is not None:
print('Displaying plot', file=sys.stderr)
title = ('Multilabel metrics with %s' %
', '.join('{0}={1}'.format(field, getattr(args, field))
for field in ['samples', 'classes', 'density']
if args.plot != field))
_plot(results, args.metrics, args.formats, title, steps, args.plot)
| bsd-3-clause |
mbayon/TFG-MachineLearning | vbig/lib/python2.7/site-packages/pandas/io/json/table_schema.py | 12 | 5184 | """
Table Schema builders
http://specs.frictionlessdata.io/json-table-schema/
"""
from pandas.core.dtypes.common import (
is_integer_dtype, is_timedelta64_dtype, is_numeric_dtype,
is_bool_dtype, is_datetime64_dtype, is_datetime64tz_dtype,
is_categorical_dtype, is_period_dtype, is_string_dtype
)
def as_json_table_type(x):
"""
Convert a NumPy / pandas type to its corresponding json_table.
Parameters
----------
x : array or dtype
Returns
-------
t : str
the Table Schema data types
Notes
-----
This table shows the relationship between NumPy / pandas dtypes,
and Table Schema dtypes.
============== =================
Pandas type Table Schema type
============== =================
int64 integer
float64 number
bool boolean
datetime64[ns] datetime
timedelta64[ns] duration
object str
categorical any
=============== =================
"""
if is_integer_dtype(x):
return 'integer'
elif is_bool_dtype(x):
return 'boolean'
elif is_numeric_dtype(x):
return 'number'
elif (is_datetime64_dtype(x) or is_datetime64tz_dtype(x) or
is_period_dtype(x)):
return 'datetime'
elif is_timedelta64_dtype(x):
return 'duration'
elif is_categorical_dtype(x):
return 'any'
elif is_string_dtype(x):
return 'string'
else:
return 'any'
def set_default_names(data):
"""Sets index names to 'index' for regular, or 'level_x' for Multi"""
if all(name is not None for name in data.index.names):
return data
data = data.copy()
if data.index.nlevels > 1:
names = [name if name is not None else 'level_{}'.format(i)
for i, name in enumerate(data.index.names)]
data.index.names = names
else:
data.index.name = data.index.name or 'index'
return data
def make_field(arr, dtype=None):
dtype = dtype or arr.dtype
if arr.name is None:
name = 'values'
else:
name = arr.name
field = {'name': name,
'type': as_json_table_type(dtype)}
if is_categorical_dtype(arr):
if hasattr(arr, 'categories'):
cats = arr.categories
ordered = arr.ordered
else:
cats = arr.cat.categories
ordered = arr.cat.ordered
field['constraints'] = {"enum": list(cats)}
field['ordered'] = ordered
elif is_period_dtype(arr):
field['freq'] = arr.freqstr
elif is_datetime64tz_dtype(arr):
if hasattr(arr, 'dt'):
field['tz'] = arr.dt.tz.zone
else:
field['tz'] = arr.tz.zone
return field
def build_table_schema(data, index=True, primary_key=None, version=True):
"""
Create a Table schema from ``data``.
Parameters
----------
data : Series, DataFrame
index : bool, default True
Whether to include ``data.index`` in the schema.
primary_key : bool or None, default True
column names to designate as the primary key.
The default `None` will set `'primaryKey'` to the index
level or levels if the index is unique.
version : bool, default True
Whether to include a field `pandas_version` with the version
of pandas that generated the schema.
Returns
-------
schema : dict
Examples
--------
>>> df = pd.DataFrame(
... {'A': [1, 2, 3],
... 'B': ['a', 'b', 'c'],
... 'C': pd.date_range('2016-01-01', freq='d', periods=3),
... }, index=pd.Index(range(3), name='idx'))
>>> build_table_schema(df)
{'fields': [{'name': 'idx', 'type': 'integer'},
{'name': 'A', 'type': 'integer'},
{'name': 'B', 'type': 'string'},
{'name': 'C', 'type': 'datetime'}],
'pandas_version': '0.20.0',
'primaryKey': ['idx']}
Notes
-----
See `_as_json_table_type` for conversion types.
Timedeltas as converted to ISO8601 duration format with
9 decimal places after the secnods field for nanosecond precision.
Categoricals are converted to the `any` dtype, and use the `enum` field
constraint to list the allowed values. The `ordered` attribute is included
in an `ordered` field.
"""
if index is True:
data = set_default_names(data)
schema = {}
fields = []
if index:
if data.index.nlevels > 1:
for level in data.index.levels:
fields.append(make_field(level))
else:
fields.append(make_field(data.index))
if data.ndim > 1:
for column, s in data.iteritems():
fields.append(make_field(s))
else:
fields.append(make_field(data))
schema['fields'] = fields
if index and data.index.is_unique and primary_key is None:
if data.index.nlevels == 1:
schema['primaryKey'] = [data.index.name]
else:
schema['primaryKey'] = data.index.names
elif primary_key is not None:
schema['primaryKey'] = primary_key
if version:
schema['pandas_version'] = '0.20.0'
return schema
| mit |
xavierwu/scikit-learn | sklearn/cluster/tests/test_k_means.py | 63 | 26190 | """Testing for K-means"""
import sys
import numpy as np
from scipy import sparse as sp
from sklearn.utils.testing import assert_equal
from sklearn.utils.testing import assert_array_equal
from sklearn.utils.testing import assert_array_almost_equal
from sklearn.utils.testing import SkipTest
from sklearn.utils.testing import assert_almost_equal
from sklearn.utils.testing import assert_raises
from sklearn.utils.testing import assert_raises_regexp
from sklearn.utils.testing import assert_true
from sklearn.utils.testing import assert_greater
from sklearn.utils.testing import assert_less
from sklearn.utils.testing import assert_warns
from sklearn.utils.testing import if_safe_multiprocessing_with_blas
from sklearn.utils.validation import DataConversionWarning
from sklearn.utils.extmath import row_norms
from sklearn.metrics.cluster import v_measure_score
from sklearn.cluster import KMeans, k_means
from sklearn.cluster import MiniBatchKMeans
from sklearn.cluster.k_means_ import _labels_inertia
from sklearn.cluster.k_means_ import _mini_batch_step
from sklearn.datasets.samples_generator import make_blobs
from sklearn.externals.six.moves import cStringIO as StringIO
# non centered, sparse centers to check the
centers = np.array([
[0.0, 5.0, 0.0, 0.0, 0.0],
[1.0, 1.0, 4.0, 0.0, 0.0],
[1.0, 0.0, 0.0, 5.0, 1.0],
])
n_samples = 100
n_clusters, n_features = centers.shape
X, true_labels = make_blobs(n_samples=n_samples, centers=centers,
cluster_std=1., random_state=42)
X_csr = sp.csr_matrix(X)
def test_kmeans_dtype():
rnd = np.random.RandomState(0)
X = rnd.normal(size=(40, 2))
X = (X * 10).astype(np.uint8)
km = KMeans(n_init=1).fit(X)
pred_x = assert_warns(DataConversionWarning, km.predict, X)
assert_array_equal(km.labels_, pred_x)
def test_labels_assignment_and_inertia():
# pure numpy implementation as easily auditable reference gold
# implementation
rng = np.random.RandomState(42)
noisy_centers = centers + rng.normal(size=centers.shape)
labels_gold = - np.ones(n_samples, dtype=np.int)
mindist = np.empty(n_samples)
mindist.fill(np.infty)
for center_id in range(n_clusters):
dist = np.sum((X - noisy_centers[center_id]) ** 2, axis=1)
labels_gold[dist < mindist] = center_id
mindist = np.minimum(dist, mindist)
inertia_gold = mindist.sum()
assert_true((mindist >= 0.0).all())
assert_true((labels_gold != -1).all())
# perform label assignment using the dense array input
x_squared_norms = (X ** 2).sum(axis=1)
labels_array, inertia_array = _labels_inertia(
X, x_squared_norms, noisy_centers)
assert_array_almost_equal(inertia_array, inertia_gold)
assert_array_equal(labels_array, labels_gold)
# perform label assignment using the sparse CSR input
x_squared_norms_from_csr = row_norms(X_csr, squared=True)
labels_csr, inertia_csr = _labels_inertia(
X_csr, x_squared_norms_from_csr, noisy_centers)
assert_array_almost_equal(inertia_csr, inertia_gold)
assert_array_equal(labels_csr, labels_gold)
def test_minibatch_update_consistency():
# Check that dense and sparse minibatch update give the same results
rng = np.random.RandomState(42)
old_centers = centers + rng.normal(size=centers.shape)
new_centers = old_centers.copy()
new_centers_csr = old_centers.copy()
counts = np.zeros(new_centers.shape[0], dtype=np.int32)
counts_csr = np.zeros(new_centers.shape[0], dtype=np.int32)
x_squared_norms = (X ** 2).sum(axis=1)
x_squared_norms_csr = row_norms(X_csr, squared=True)
buffer = np.zeros(centers.shape[1], dtype=np.double)
buffer_csr = np.zeros(centers.shape[1], dtype=np.double)
# extract a small minibatch
X_mb = X[:10]
X_mb_csr = X_csr[:10]
x_mb_squared_norms = x_squared_norms[:10]
x_mb_squared_norms_csr = x_squared_norms_csr[:10]
# step 1: compute the dense minibatch update
old_inertia, incremental_diff = _mini_batch_step(
X_mb, x_mb_squared_norms, new_centers, counts,
buffer, 1, None, random_reassign=False)
assert_greater(old_inertia, 0.0)
# compute the new inertia on the same batch to check that it decreased
labels, new_inertia = _labels_inertia(
X_mb, x_mb_squared_norms, new_centers)
assert_greater(new_inertia, 0.0)
assert_less(new_inertia, old_inertia)
# check that the incremental difference computation is matching the
# final observed value
effective_diff = np.sum((new_centers - old_centers) ** 2)
assert_almost_equal(incremental_diff, effective_diff)
# step 2: compute the sparse minibatch update
old_inertia_csr, incremental_diff_csr = _mini_batch_step(
X_mb_csr, x_mb_squared_norms_csr, new_centers_csr, counts_csr,
buffer_csr, 1, None, random_reassign=False)
assert_greater(old_inertia_csr, 0.0)
# compute the new inertia on the same batch to check that it decreased
labels_csr, new_inertia_csr = _labels_inertia(
X_mb_csr, x_mb_squared_norms_csr, new_centers_csr)
assert_greater(new_inertia_csr, 0.0)
assert_less(new_inertia_csr, old_inertia_csr)
# check that the incremental difference computation is matching the
# final observed value
effective_diff = np.sum((new_centers_csr - old_centers) ** 2)
assert_almost_equal(incremental_diff_csr, effective_diff)
# step 3: check that sparse and dense updates lead to the same results
assert_array_equal(labels, labels_csr)
assert_array_almost_equal(new_centers, new_centers_csr)
assert_almost_equal(incremental_diff, incremental_diff_csr)
assert_almost_equal(old_inertia, old_inertia_csr)
assert_almost_equal(new_inertia, new_inertia_csr)
def _check_fitted_model(km):
# check that the number of clusters centers and distinct labels match
# the expectation
centers = km.cluster_centers_
assert_equal(centers.shape, (n_clusters, n_features))
labels = km.labels_
assert_equal(np.unique(labels).shape[0], n_clusters)
# check that the labels assignment are perfect (up to a permutation)
assert_equal(v_measure_score(true_labels, labels), 1.0)
assert_greater(km.inertia_, 0.0)
# check error on dataset being too small
assert_raises(ValueError, km.fit, [[0., 1.]])
def test_k_means_plus_plus_init():
km = KMeans(init="k-means++", n_clusters=n_clusters,
random_state=42).fit(X)
_check_fitted_model(km)
def test_k_means_new_centers():
# Explore the part of the code where a new center is reassigned
X = np.array([[0, 0, 1, 1],
[0, 0, 0, 0],
[0, 1, 0, 0],
[0, 0, 0, 0],
[0, 0, 0, 0],
[0, 1, 0, 0]])
labels = [0, 1, 2, 1, 1, 2]
bad_centers = np.array([[+0, 1, 0, 0],
[.2, 0, .2, .2],
[+0, 0, 0, 0]])
km = KMeans(n_clusters=3, init=bad_centers, n_init=1, max_iter=10,
random_state=1)
for this_X in (X, sp.coo_matrix(X)):
km.fit(this_X)
this_labels = km.labels_
# Reorder the labels so that the first instance is in cluster 0,
# the second in cluster 1, ...
this_labels = np.unique(this_labels, return_index=True)[1][this_labels]
np.testing.assert_array_equal(this_labels, labels)
@if_safe_multiprocessing_with_blas
def test_k_means_plus_plus_init_2_jobs():
if sys.version_info[:2] < (3, 4):
raise SkipTest(
"Possible multi-process bug with some BLAS under Python < 3.4")
km = KMeans(init="k-means++", n_clusters=n_clusters, n_jobs=2,
random_state=42).fit(X)
_check_fitted_model(km)
def test_k_means_precompute_distances_flag():
# check that a warning is raised if the precompute_distances flag is not
# supported
km = KMeans(precompute_distances="wrong")
assert_raises(ValueError, km.fit, X)
def test_k_means_plus_plus_init_sparse():
km = KMeans(init="k-means++", n_clusters=n_clusters, random_state=42)
km.fit(X_csr)
_check_fitted_model(km)
def test_k_means_random_init():
km = KMeans(init="random", n_clusters=n_clusters, random_state=42)
km.fit(X)
_check_fitted_model(km)
def test_k_means_random_init_sparse():
km = KMeans(init="random", n_clusters=n_clusters, random_state=42)
km.fit(X_csr)
_check_fitted_model(km)
def test_k_means_plus_plus_init_not_precomputed():
km = KMeans(init="k-means++", n_clusters=n_clusters, random_state=42,
precompute_distances=False).fit(X)
_check_fitted_model(km)
def test_k_means_random_init_not_precomputed():
km = KMeans(init="random", n_clusters=n_clusters, random_state=42,
precompute_distances=False).fit(X)
_check_fitted_model(km)
def test_k_means_perfect_init():
km = KMeans(init=centers.copy(), n_clusters=n_clusters, random_state=42,
n_init=1)
km.fit(X)
_check_fitted_model(km)
def test_k_means_n_init():
rnd = np.random.RandomState(0)
X = rnd.normal(size=(40, 2))
# two regression tests on bad n_init argument
# previous bug: n_init <= 0 threw non-informative TypeError (#3858)
assert_raises_regexp(ValueError, "n_init", KMeans(n_init=0).fit, X)
assert_raises_regexp(ValueError, "n_init", KMeans(n_init=-1).fit, X)
def test_k_means_fortran_aligned_data():
# Check the KMeans will work well, even if X is a fortran-aligned data.
X = np.asfortranarray([[0, 0], [0, 1], [0, 1]])
centers = np.array([[0, 0], [0, 1]])
labels = np.array([0, 1, 1])
km = KMeans(n_init=1, init=centers, precompute_distances=False,
random_state=42)
km.fit(X)
assert_array_equal(km.cluster_centers_, centers)
assert_array_equal(km.labels_, labels)
def test_mb_k_means_plus_plus_init_dense_array():
mb_k_means = MiniBatchKMeans(init="k-means++", n_clusters=n_clusters,
random_state=42)
mb_k_means.fit(X)
_check_fitted_model(mb_k_means)
def test_mb_kmeans_verbose():
mb_k_means = MiniBatchKMeans(init="k-means++", n_clusters=n_clusters,
random_state=42, verbose=1)
old_stdout = sys.stdout
sys.stdout = StringIO()
try:
mb_k_means.fit(X)
finally:
sys.stdout = old_stdout
def test_mb_k_means_plus_plus_init_sparse_matrix():
mb_k_means = MiniBatchKMeans(init="k-means++", n_clusters=n_clusters,
random_state=42)
mb_k_means.fit(X_csr)
_check_fitted_model(mb_k_means)
def test_minibatch_init_with_large_k():
mb_k_means = MiniBatchKMeans(init='k-means++', init_size=10, n_clusters=20)
# Check that a warning is raised, as the number clusters is larger
# than the init_size
assert_warns(RuntimeWarning, mb_k_means.fit, X)
def test_minibatch_k_means_random_init_dense_array():
# increase n_init to make random init stable enough
mb_k_means = MiniBatchKMeans(init="random", n_clusters=n_clusters,
random_state=42, n_init=10).fit(X)
_check_fitted_model(mb_k_means)
def test_minibatch_k_means_random_init_sparse_csr():
# increase n_init to make random init stable enough
mb_k_means = MiniBatchKMeans(init="random", n_clusters=n_clusters,
random_state=42, n_init=10).fit(X_csr)
_check_fitted_model(mb_k_means)
def test_minibatch_k_means_perfect_init_dense_array():
mb_k_means = MiniBatchKMeans(init=centers.copy(), n_clusters=n_clusters,
random_state=42, n_init=1).fit(X)
_check_fitted_model(mb_k_means)
def test_minibatch_k_means_init_multiple_runs_with_explicit_centers():
mb_k_means = MiniBatchKMeans(init=centers.copy(), n_clusters=n_clusters,
random_state=42, n_init=10)
assert_warns(RuntimeWarning, mb_k_means.fit, X)
def test_minibatch_k_means_perfect_init_sparse_csr():
mb_k_means = MiniBatchKMeans(init=centers.copy(), n_clusters=n_clusters,
random_state=42, n_init=1).fit(X_csr)
_check_fitted_model(mb_k_means)
def test_minibatch_sensible_reassign_fit():
# check if identical initial clusters are reassigned
# also a regression test for when there are more desired reassignments than
# samples.
zeroed_X, true_labels = make_blobs(n_samples=100, centers=5,
cluster_std=1., random_state=42)
zeroed_X[::2, :] = 0
mb_k_means = MiniBatchKMeans(n_clusters=20, batch_size=10, random_state=42,
init="random")
mb_k_means.fit(zeroed_X)
# there should not be too many exact zero cluster centers
assert_greater(mb_k_means.cluster_centers_.any(axis=1).sum(), 10)
# do the same with batch-size > X.shape[0] (regression test)
mb_k_means = MiniBatchKMeans(n_clusters=20, batch_size=201,
random_state=42, init="random")
mb_k_means.fit(zeroed_X)
# there should not be too many exact zero cluster centers
assert_greater(mb_k_means.cluster_centers_.any(axis=1).sum(), 10)
def test_minibatch_sensible_reassign_partial_fit():
zeroed_X, true_labels = make_blobs(n_samples=n_samples, centers=5,
cluster_std=1., random_state=42)
zeroed_X[::2, :] = 0
mb_k_means = MiniBatchKMeans(n_clusters=20, random_state=42, init="random")
for i in range(100):
mb_k_means.partial_fit(zeroed_X)
# there should not be too many exact zero cluster centers
assert_greater(mb_k_means.cluster_centers_.any(axis=1).sum(), 10)
def test_minibatch_reassign():
# Give a perfect initialization, but a large reassignment_ratio,
# as a result all the centers should be reassigned and the model
# should not longer be good
for this_X in (X, X_csr):
mb_k_means = MiniBatchKMeans(n_clusters=n_clusters, batch_size=100,
random_state=42)
mb_k_means.fit(this_X)
score_before = mb_k_means.score(this_X)
try:
old_stdout = sys.stdout
sys.stdout = StringIO()
# Turn on verbosity to smoke test the display code
_mini_batch_step(this_X, (X ** 2).sum(axis=1),
mb_k_means.cluster_centers_,
mb_k_means.counts_,
np.zeros(X.shape[1], np.double),
False, distances=np.zeros(X.shape[0]),
random_reassign=True, random_state=42,
reassignment_ratio=1, verbose=True)
finally:
sys.stdout = old_stdout
assert_greater(score_before, mb_k_means.score(this_X))
# Give a perfect initialization, with a small reassignment_ratio,
# no center should be reassigned
for this_X in (X, X_csr):
mb_k_means = MiniBatchKMeans(n_clusters=n_clusters, batch_size=100,
init=centers.copy(),
random_state=42, n_init=1)
mb_k_means.fit(this_X)
clusters_before = mb_k_means.cluster_centers_
# Turn on verbosity to smoke test the display code
_mini_batch_step(this_X, (X ** 2).sum(axis=1),
mb_k_means.cluster_centers_,
mb_k_means.counts_,
np.zeros(X.shape[1], np.double),
False, distances=np.zeros(X.shape[0]),
random_reassign=True, random_state=42,
reassignment_ratio=1e-15)
assert_array_almost_equal(clusters_before, mb_k_means.cluster_centers_)
def test_minibatch_with_many_reassignments():
# Test for the case that the number of clusters to reassign is bigger
# than the batch_size
n_samples = 550
rnd = np.random.RandomState(42)
X = rnd.uniform(size=(n_samples, 10))
# Check that the fit works if n_clusters is bigger than the batch_size.
# Run the test with 550 clusters and 550 samples, because it turned out
# that this values ensure that the number of clusters to reassign
# is always bigger than the batch_size
n_clusters = 550
MiniBatchKMeans(n_clusters=n_clusters,
batch_size=100,
init_size=n_samples,
random_state=42).fit(X)
def test_sparse_mb_k_means_callable_init():
def test_init(X, k, random_state):
return centers
# Small test to check that giving the wrong number of centers
# raises a meaningful error
assert_raises(ValueError,
MiniBatchKMeans(init=test_init, random_state=42).fit, X_csr)
# Now check that the fit actually works
mb_k_means = MiniBatchKMeans(n_clusters=3, init=test_init,
random_state=42).fit(X_csr)
_check_fitted_model(mb_k_means)
def test_mini_batch_k_means_random_init_partial_fit():
km = MiniBatchKMeans(n_clusters=n_clusters, init="random", random_state=42)
# use the partial_fit API for online learning
for X_minibatch in np.array_split(X, 10):
km.partial_fit(X_minibatch)
# compute the labeling on the complete dataset
labels = km.predict(X)
assert_equal(v_measure_score(true_labels, labels), 1.0)
def test_minibatch_default_init_size():
mb_k_means = MiniBatchKMeans(init=centers.copy(), n_clusters=n_clusters,
batch_size=10, random_state=42,
n_init=1).fit(X)
assert_equal(mb_k_means.init_size_, 3 * mb_k_means.batch_size)
_check_fitted_model(mb_k_means)
def test_minibatch_tol():
mb_k_means = MiniBatchKMeans(n_clusters=n_clusters, batch_size=10,
random_state=42, tol=.01).fit(X)
_check_fitted_model(mb_k_means)
def test_minibatch_set_init_size():
mb_k_means = MiniBatchKMeans(init=centers.copy(), n_clusters=n_clusters,
init_size=666, random_state=42,
n_init=1).fit(X)
assert_equal(mb_k_means.init_size, 666)
assert_equal(mb_k_means.init_size_, n_samples)
_check_fitted_model(mb_k_means)
def test_k_means_invalid_init():
km = KMeans(init="invalid", n_init=1, n_clusters=n_clusters)
assert_raises(ValueError, km.fit, X)
def test_mini_match_k_means_invalid_init():
km = MiniBatchKMeans(init="invalid", n_init=1, n_clusters=n_clusters)
assert_raises(ValueError, km.fit, X)
def test_k_means_copyx():
# Check if copy_x=False returns nearly equal X after de-centering.
my_X = X.copy()
km = KMeans(copy_x=False, n_clusters=n_clusters, random_state=42)
km.fit(my_X)
_check_fitted_model(km)
# check if my_X is centered
assert_array_almost_equal(my_X, X)
def test_k_means_non_collapsed():
# Check k_means with a bad initialization does not yield a singleton
# Starting with bad centers that are quickly ignored should not
# result in a repositioning of the centers to the center of mass that
# would lead to collapsed centers which in turns make the clustering
# dependent of the numerical unstabilities.
my_X = np.array([[1.1, 1.1], [0.9, 1.1], [1.1, 0.9], [0.9, 1.1]])
array_init = np.array([[1.0, 1.0], [5.0, 5.0], [-5.0, -5.0]])
km = KMeans(init=array_init, n_clusters=3, random_state=42, n_init=1)
km.fit(my_X)
# centers must not been collapsed
assert_equal(len(np.unique(km.labels_)), 3)
centers = km.cluster_centers_
assert_true(np.linalg.norm(centers[0] - centers[1]) >= 0.1)
assert_true(np.linalg.norm(centers[0] - centers[2]) >= 0.1)
assert_true(np.linalg.norm(centers[1] - centers[2]) >= 0.1)
def test_predict():
km = KMeans(n_clusters=n_clusters, random_state=42)
km.fit(X)
# sanity check: predict centroid labels
pred = km.predict(km.cluster_centers_)
assert_array_equal(pred, np.arange(n_clusters))
# sanity check: re-predict labeling for training set samples
pred = km.predict(X)
assert_array_equal(pred, km.labels_)
# re-predict labels for training set using fit_predict
pred = km.fit_predict(X)
assert_array_equal(pred, km.labels_)
def test_score():
km1 = KMeans(n_clusters=n_clusters, max_iter=1, random_state=42)
s1 = km1.fit(X).score(X)
km2 = KMeans(n_clusters=n_clusters, max_iter=10, random_state=42)
s2 = km2.fit(X).score(X)
assert_greater(s2, s1)
def test_predict_minibatch_dense_input():
mb_k_means = MiniBatchKMeans(n_clusters=n_clusters, random_state=40).fit(X)
# sanity check: predict centroid labels
pred = mb_k_means.predict(mb_k_means.cluster_centers_)
assert_array_equal(pred, np.arange(n_clusters))
# sanity check: re-predict labeling for training set samples
pred = mb_k_means.predict(X)
assert_array_equal(mb_k_means.predict(X), mb_k_means.labels_)
def test_predict_minibatch_kmeanspp_init_sparse_input():
mb_k_means = MiniBatchKMeans(n_clusters=n_clusters, init='k-means++',
n_init=10).fit(X_csr)
# sanity check: re-predict labeling for training set samples
assert_array_equal(mb_k_means.predict(X_csr), mb_k_means.labels_)
# sanity check: predict centroid labels
pred = mb_k_means.predict(mb_k_means.cluster_centers_)
assert_array_equal(pred, np.arange(n_clusters))
# check that models trained on sparse input also works for dense input at
# predict time
assert_array_equal(mb_k_means.predict(X), mb_k_means.labels_)
def test_predict_minibatch_random_init_sparse_input():
mb_k_means = MiniBatchKMeans(n_clusters=n_clusters, init='random',
n_init=10).fit(X_csr)
# sanity check: re-predict labeling for training set samples
assert_array_equal(mb_k_means.predict(X_csr), mb_k_means.labels_)
# sanity check: predict centroid labels
pred = mb_k_means.predict(mb_k_means.cluster_centers_)
assert_array_equal(pred, np.arange(n_clusters))
# check that models trained on sparse input also works for dense input at
# predict time
assert_array_equal(mb_k_means.predict(X), mb_k_means.labels_)
def test_input_dtypes():
X_list = [[0, 0], [10, 10], [12, 9], [-1, 1], [2, 0], [8, 10]]
X_int = np.array(X_list, dtype=np.int32)
X_int_csr = sp.csr_matrix(X_int)
init_int = X_int[:2]
fitted_models = [
KMeans(n_clusters=2).fit(X_list),
KMeans(n_clusters=2).fit(X_int),
KMeans(n_clusters=2, init=init_int, n_init=1).fit(X_list),
KMeans(n_clusters=2, init=init_int, n_init=1).fit(X_int),
# mini batch kmeans is very unstable on such a small dataset hence
# we use many inits
MiniBatchKMeans(n_clusters=2, n_init=10, batch_size=2).fit(X_list),
MiniBatchKMeans(n_clusters=2, n_init=10, batch_size=2).fit(X_int),
MiniBatchKMeans(n_clusters=2, n_init=10, batch_size=2).fit(X_int_csr),
MiniBatchKMeans(n_clusters=2, batch_size=2,
init=init_int, n_init=1).fit(X_list),
MiniBatchKMeans(n_clusters=2, batch_size=2,
init=init_int, n_init=1).fit(X_int),
MiniBatchKMeans(n_clusters=2, batch_size=2,
init=init_int, n_init=1).fit(X_int_csr),
]
expected_labels = [0, 1, 1, 0, 0, 1]
scores = np.array([v_measure_score(expected_labels, km.labels_)
for km in fitted_models])
assert_array_equal(scores, np.ones(scores.shape[0]))
def test_transform():
km = KMeans(n_clusters=n_clusters)
km.fit(X)
X_new = km.transform(km.cluster_centers_)
for c in range(n_clusters):
assert_equal(X_new[c, c], 0)
for c2 in range(n_clusters):
if c != c2:
assert_greater(X_new[c, c2], 0)
def test_fit_transform():
X1 = KMeans(n_clusters=3, random_state=51).fit(X).transform(X)
X2 = KMeans(n_clusters=3, random_state=51).fit_transform(X)
assert_array_equal(X1, X2)
def test_n_init():
# Check that increasing the number of init increases the quality
n_runs = 5
n_init_range = [1, 5, 10]
inertia = np.zeros((len(n_init_range), n_runs))
for i, n_init in enumerate(n_init_range):
for j in range(n_runs):
km = KMeans(n_clusters=n_clusters, init="random", n_init=n_init,
random_state=j).fit(X)
inertia[i, j] = km.inertia_
inertia = inertia.mean(axis=1)
failure_msg = ("Inertia %r should be decreasing"
" when n_init is increasing.") % list(inertia)
for i in range(len(n_init_range) - 1):
assert_true(inertia[i] >= inertia[i + 1], failure_msg)
def test_k_means_function():
# test calling the k_means function directly
# catch output
old_stdout = sys.stdout
sys.stdout = StringIO()
try:
cluster_centers, labels, inertia = k_means(X, n_clusters=n_clusters,
verbose=True)
finally:
sys.stdout = old_stdout
centers = cluster_centers
assert_equal(centers.shape, (n_clusters, n_features))
labels = labels
assert_equal(np.unique(labels).shape[0], n_clusters)
# check that the labels assignment are perfect (up to a permutation)
assert_equal(v_measure_score(true_labels, labels), 1.0)
assert_greater(inertia, 0.0)
# check warning when centers are passed
assert_warns(RuntimeWarning, k_means, X, n_clusters=n_clusters,
init=centers)
# to many clusters desired
assert_raises(ValueError, k_means, X, n_clusters=X.shape[0] + 1)
def test_x_squared_norms_init_centroids():
"""Test that x_squared_norms can be None in _init_centroids"""
from sklearn.cluster.k_means_ import _init_centroids
X_norms = np.sum(X**2, axis=1)
precompute = _init_centroids(
X, 3, "k-means++", random_state=0, x_squared_norms=X_norms)
assert_array_equal(
precompute,
_init_centroids(X, 3, "k-means++", random_state=0))
| bsd-3-clause |
JensTimmerman/radical.pilot | src/radical/pilot/utils/analysis.py | 1 | 12671 |
import os
# ------------------------------------------------------------------------------
#
def get_experiment_frames(experiments, datadir=None):
"""
read profiles for all sessions in the given 'experiments' dict. That dict
is expected to be like this:
{ 'test 1' : [ [ 'rp.session.thinkie.merzky.016609.0007', 'stampede popen sleep 1/1/1/1 (?)'] ],
'test 2' : [ [ 'rp.session.ip-10-184-31-85.merzky.016610.0112', 'stampede shell sleep 16/8/8/4' ] ],
'test 3' : [ [ 'rp.session.ip-10-184-31-85.merzky.016611.0013', 'stampede shell mdrun 16/8/8/4' ] ],
'test 4' : [ [ 'rp.session.titan-ext4.marksant1.016607.0005', 'titan shell sleep 1/1/1/1 a' ] ],
'test 5' : [ [ 'rp.session.titan-ext4.marksant1.016607.0006', 'titan shell sleep 1/1/1/1 b' ] ],
'test 6' : [ [ 'rp.session.ip-10-184-31-85.merzky.016611.0013', 'stampede - isolated', ],
[ 'rp.session.ip-10-184-31-85.merzky.016612.0012', 'stampede - integrated', ],
[ 'rp.session.titan-ext4.marksant1.016607.0006', 'blue waters - integrated' ] ]
} name in
ie. iname in t is a list of experiment names, and each label has a list of
session/label pairs, where the label will be later used to label (duh) plots.
we return a similar dict where the session IDs are data frames
"""
import pandas as pd
exp_frames = dict()
if not datadir:
datadir = os.getcwd()
print 'reading profiles in %s' % datadir
for exp in experiments:
print " - %s" % exp
exp_frames[exp] = list()
for sid, label in experiments[exp]:
print " - %s" % sid
import glob
for prof in glob.glob ("%s/%s-pilot.*.prof" % (datadir, sid)):
print " - %s" % prof
frame = get_profile_frame (prof)
exp_frames[exp].append ([frame, label])
return exp_frames
# ------------------------------------------------------------------------------
#
def get_profile_frame (prof):
import pandas as pd
return pd.read_csv(prof)
# ------------------------------------------------------------------------------
#
tmp = None
def add_concurrency (frame, tgt, spec):
"""
add a column 'tgt' which is a cumulative sum of conditionals of enother row.
The purpose is the following: if a unit enters a component, the tgt row counter is
increased by 1, if the unit leaves the component, the counter is decreases by 1.
For any time, the resulting row contains the number of units which is in the
component. Or state. Or whatever.
The arguments are:
'tgt' : name of the new column
'spec' : a set of filters to determine if a unit enters or leaves
'spec' is expected to be a dict of the following format:
spec = { 'in' : [{'col1' : 'pat1',
'col2' : 'pat2'},
...],
'out' : [{'col3' : 'pat3',
'col4' : 'pat4'},
...]
}
where:
'in' : filter set to determine the unit entering
'out' : filter set to determine the unit leaving
'col' : name of column for which filter is defined
'event' : event which correlates to entering/leaving
'msg' : qualifier on the event, if event is not unique
Example:
spec = {'in' : [{'state' :'Executing'}],
'out' : [{'state' :'Done'},
{'state' :'Failed'},
{'state' :'Cancelled'}]
}
get_concurrency (df, 'concurrently_running', spec)
"""
import numpy
# create a temporary row over which we can do the commulative sum
# --------------------------------------------------------------------------
def _conc (row, spec):
# row must match any filter dict in 'spec[in/out]'
# for any filter dict it must match all col/pat pairs
# for each in filter
for f in spec['in']:
match = 1
# for each col/val in that filter
for col, pat in f.iteritems():
if row[col] != pat:
match = 0
break
if match:
# one filter matched!
# print " + : %-20s : %.2f : %-20s : %s " % (row['uid'], row['time'], row['event'], row['message'])
return 1
# for each out filter
for f in spec['out']:
match = 1
# for each col/val in that filter
for col, pat in f.iteritems():
if row[col] != pat:
match = 0
break
if match:
# one filter matched!
# print " - : %-20s : %.2f : %-20s : %s " % (row['uid'], row['time'], row['event'], row['message'])
return -1
# no filter matched
# print " : %-20s : %.2f : %-20s : %s " % (row['uid'], row['time'], row['event'], row['message'])
return 0
# --------------------------------------------------------------------------
# we only want to later look at changes of the concurrency -- leading or trailing
# idle times are to be ignored. We thus set repeating values of the cumsum to NaN,
# so that they can be filtered out when ploting: df.dropna().plot(...).
# That specifically will limit the plotted time range to the area of activity.
# The full time range can still be plotted when ommitting the dropna() call.
# --------------------------------------------------------------------------
def _time (x):
global tmp
if x != tmp: tmp = x
else : x = numpy.NaN
return x
# --------------------------------------------------------------------------
# sanitize concurrency: negative values indicate incorrect event ordering,
# so we set the repesctive values to 0
# --------------------------------------------------------------------------
def _abs (x):
if x < 0:
return numpy.NaN
return x
# --------------------------------------------------------------------------
frame[tgt] = frame.apply(lambda row: _conc(row, spec), axis=1).cumsum()
frame[tgt] = frame.apply(lambda row: _abs (row[tgt]), axis=1)
frame[tgt] = frame.apply(lambda row: _time(row[tgt]), axis=1)
# print frame[[tgt, 'time']]
# ------------------------------------------------------------------------------
#
t0 = None
def calibrate_frame(frame, spec):
"""
move the time axis of a profiling frame so that t_0 is at the first event
matching the given 'spec'. 'spec' has the same format as described in
'add_concurrency' (list of dicts with col:pat filters)
"""
# --------------------------------------------------------------------------
def _find_t0 (row, spec):
# row must match any filter dict in 'spec[in/out]'
# for any filter dict it must match all col/pat pairs
global t0
if t0 is not None:
# already found t0
return
# for each col/val in that filter
for f in spec:
match = 1
for col, pat in f.iteritems():
if row[col] != pat:
match = 0
break
if match:
# one filter matched!
t0 = row['time']
return
# --------------------------------------------------------------------------
# --------------------------------------------------------------------------
def _calibrate (row, t0):
if t0 is None:
# no t0...
return
return row['time'] - t0
# --------------------------------------------------------------------------
# we need to iterate twice over the frame: first to find t0, then to
# calibrate the time axis
global t0
t0 = None # no t0
frame.apply(lambda row: _find_t0 (row, spec), axis=1)
if t0 == None:
print "Can't recalibrate, no matching timestamp found"
return
frame['time'] = frame.apply(lambda row: _calibrate(row, t0 ), axis=1)
# ------------------------------------------------------------------------------
#
def create_plot():
"""
create a plot object and tune its layout to our liking.
"""
import matplotlib.pyplot as plt
fig, plot = plt.subplots(figsize=(12,6))
plot.xaxis.set_tick_params(width=1, length=7)
plot.yaxis.set_tick_params(width=1, length=7)
plot.spines['right' ].set_position(('outward', 10))
plot.spines['top' ].set_position(('outward', 10))
plot.spines['bottom'].set_position(('outward', 10))
plot.spines['left' ].set_position(('outward', 10))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
fig.tight_layout()
return fig, plot
# ------------------------------------------------------------------------------
#
def frame_plot (frames, axis, title=None, logx=False, logy=False,
legend=True, figdir=None):
"""
plot the given axis from the give data frame. We create a plot, and plot
all frames given in the list. The list is expected to contain [frame,label]
pairs
frames: list of tuples of dataframes and labels
frames = [[stampede_df_1, 'stampede - popen'],
[stampede_df_2, 'stampede - shell'],
[stampede_df_3, 'stampede - ORTE' ]]
axis: tuple of data frame column index and axis label
axis = ['time', 'time (s)']
"""
# create figure and layout
fig, plot = create_plot()
# set plot title
if title:
plot.set_title(title, y=1.05, fontsize=18)
# plot the data frames
# NOTE: we need to set labels separately, because of
# https://github.com/pydata/pandas/issues/9542
labels = list()
for frame, label in frames:
try:
frame.dropna().plot(ax=plot, logx=logx, logy=logy,
x=axis[0][0], y=axis[1][0],
drawstyle='steps',
label=label, legend=False)
except Exception as e:
print "skipping frame '%s': '%s'" % (label, e)
if legend:
plot.legend(labels=labels, loc='upper right', fontsize=14, frameon=True)
# set axis labels
plot.set_xlabel(axis[0][1], fontsize=14)
plot.set_ylabel(axis[1][1], fontsize=14)
plot.set_frame_on(True)
# save as png and pdf. Use the title as base for names
if title: base = title
else : base = "%s_%s" % (axis[0][1], axis[1][1])
# clean up base name -- only keep alphanum and such
import re
base = re.sub('[^a-zA-Z0-9\.\-]', '_', base)
base = re.sub('_+', '_', base)
if not figdir:
figdir = os.getcwd()
print 'saving %s/%s.png' % (figdir, base)
fig.savefig('%s/%s.png' % (figdir, base), bbox_inches='tight')
print 'saving %s/%s.pdf' % (figdir, base)
fig.savefig('%s/%s.pdf' % (figdir, base), bbox_inches='tight')
return fig, plot
# ------------------------------------------------------------------------------
#
def create_analytical_frame (idx, kind, args, limits, step):
"""
create an artificial data frame, ie. a data frame which does not contain
data gathered from an experiment, but data representing an analytical
construct of some 'kind'.
idx: data frame column index to fill (a time column is always created)
kind: construct to use (only 'rate' is supporte right now)
args: construct specific parameters
limits: time range for which data are to be created
step: time steps for which data are to be created
"""
import pandas as pd
# --------------------------------------------------------------------------
def _frange(start, stop, step):
while start <= stop:
yield start
start += step
# --------------------------------------------------------------------------
if kind == 'rate' :
t_0 = args.get ('t_0', 0.0)
rate = args.get ('rate', 1.0)
data = list()
for t in _frange(limits[0], limits[1], step):
data.append ({'time': t+t_0, idx: t*rate})
return pd.DataFrame (data)
else:
raise ValueError ("No such frame kind '%s'" % kind)
# ------------------------------------------------------------------------------
| mit |
alexandrebarachant/mne-python | mne/decoding/tests/test_ems.py | 1 | 3384 | # Author: Denis A. Engemann <d.engemann@gmail.com>
#
# License: BSD (3-clause)
import os.path as op
import numpy as np
from numpy.testing import assert_array_almost_equal
from nose.tools import assert_equal, assert_raises
from mne import io, Epochs, read_events, pick_types
from mne.utils import requires_sklearn, check_version
from mne.decoding import compute_ems, EMS
data_dir = op.join(op.dirname(__file__), '..', '..', 'io', 'tests', 'data')
curdir = op.join(op.dirname(__file__))
raw_fname = op.join(data_dir, 'test_raw.fif')
event_name = op.join(data_dir, 'test-eve.fif')
tmin, tmax = -0.2, 0.5
event_id = dict(aud_l=1, vis_l=3)
@requires_sklearn
def test_ems():
"""Test event-matched spatial filters"""
raw = io.read_raw_fif(raw_fname, preload=False)
# create unequal number of events
events = read_events(event_name)
events[-2, 2] = 3
picks = pick_types(raw.info, meg=True, stim=False, ecg=False,
eog=False, exclude='bads')
picks = picks[1:13:3]
epochs = Epochs(raw, events, event_id, tmin, tmax, picks=picks,
baseline=(None, 0), preload=True)
assert_raises(ValueError, compute_ems, epochs, ['aud_l', 'vis_l'])
epochs = epochs.equalize_event_counts(epochs.event_id, copy=False)[0]
assert_raises(KeyError, compute_ems, epochs, ['blah', 'hahah'])
surrogates, filters, conditions = compute_ems(epochs)
assert_equal(list(set(conditions)), [1, 3])
events = read_events(event_name)
event_id2 = dict(aud_l=1, aud_r=2, vis_l=3)
epochs = Epochs(raw, events, event_id2, tmin, tmax, picks=picks,
baseline=(None, 0), preload=True)
epochs = epochs.equalize_event_counts(epochs.event_id, copy=False)[0]
n_expected = sum([len(epochs[k]) for k in ['aud_l', 'vis_l']])
assert_raises(ValueError, compute_ems, epochs)
surrogates, filters, conditions = compute_ems(epochs, ['aud_r', 'vis_l'])
assert_equal(n_expected, len(surrogates))
assert_equal(n_expected, len(conditions))
assert_equal(list(set(conditions)), [2, 3])
# test compute_ems cv
epochs = epochs['aud_r', 'vis_l']
epochs.equalize_event_counts(epochs.event_id)
if check_version('sklearn', '0.18'):
from sklearn.model_selection import StratifiedKFold
cv = StratifiedKFold()
else:
from sklearn.cross_validation import StratifiedKFold
cv = StratifiedKFold(epochs.events[:, 2])
compute_ems(epochs, cv=cv)
compute_ems(epochs, cv=2)
assert_raises(ValueError, compute_ems, epochs, cv='foo')
assert_raises(ValueError, compute_ems, epochs, cv=len(epochs) + 1)
raw.close()
# EMS transformer, check that identical to compute_ems
X = epochs.get_data()
y = epochs.events[:, 2]
X = X / np.std(X) # X scaled outside cv in compute_ems
Xt, coefs = list(), list()
ems = EMS()
assert_equal(ems.__repr__(), '<EMS: not fitted.>')
# manual leave-one-out to avoid sklearn version problem
for test in range(len(y)):
train = np.setdiff1d(range(len(y)), test)
ems.fit(X[train], y[train])
coefs.append(ems.filters_)
Xt.append(ems.transform(X[[test]]))
assert_equal(ems.__repr__(), '<EMS: fitted with 4 filters on 2 classes.>')
assert_array_almost_equal(filters, np.mean(coefs, axis=0))
assert_array_almost_equal(surrogates, np.vstack(Xt))
| bsd-3-clause |
muku42/bokeh | bokeh/charts/builder/tests/test_dot_builder.py | 4 | 3924 | """ This is the Bokeh charts testing interface.
"""
#-----------------------------------------------------------------------------
# Copyright (c) 2012 - 2014, Continuum Analytics, Inc. All rights reserved.
#
# Powered by the Bokeh Development Team.
#
# The full license is in the file LICENSE.txt, distributed with this software.
#-----------------------------------------------------------------------------
#-----------------------------------------------------------------------------
# Imports
#-----------------------------------------------------------------------------
from __future__ import absolute_import
from collections import OrderedDict
import unittest
import numpy as np
from numpy.testing import assert_array_equal
import pandas as pd
from bokeh.charts import Dot
from bokeh.util.testing import create_chart
#-----------------------------------------------------------------------------
# Classes and functions
#-----------------------------------------------------------------------------
class TestDot(unittest.TestCase):
def test_supported_input(self):
xyvalues = OrderedDict()
xyvalues['python']=[2, 5]
xyvalues['pypy']=[12, 40]
xyvalues['jython']=[22, 30]
xyvaluesdf = pd.DataFrame(xyvalues, index=['lists', 'loops'])
cat = ['lists', 'loops']
catjython = ['lists:0.75', 'loops:0.75']
catpypy = ['lists:0.5', 'loops:0.5']
catpython = ['lists:0.25', 'loops:0.25']
python = seg_top_python = [2, 5]
pypy = seg_top_pypy = [12, 40]
jython = seg_top_jython = [22, 30]
zero = [0, 0]
for i, _xy in enumerate([xyvalues, xyvaluesdf]):
hm = create_chart(Dot, _xy, cat=cat)
builder = hm._builders[0]
self.assertEqual(sorted(builder._groups), sorted(list(xyvalues.keys())))
assert_array_equal(builder._data['cat'], cat)
assert_array_equal(builder._data['catjython'], catjython)
assert_array_equal(builder._data['catpython'], catpython)
assert_array_equal(builder._data['catpypy'], catpypy)
assert_array_equal(builder._data['python'], python)
assert_array_equal(builder._data['jython'], jython)
assert_array_equal(builder._data['pypy'], pypy)
assert_array_equal(builder._data['seg_top_python'], seg_top_python)
assert_array_equal(builder._data['seg_top_jython'], seg_top_jython)
assert_array_equal(builder._data['seg_top_pypy'], seg_top_pypy)
assert_array_equal(builder._data['z_python'], zero)
assert_array_equal(builder._data['z_pypy'], zero)
assert_array_equal(builder._data['z_jython'], zero)
assert_array_equal(builder._data['zero'], zero)
lvalues = [[2, 5], [12, 40], [22, 30]]
for _xy in [lvalues, np.array(lvalues)]:
hm = create_chart(Dot, _xy, cat=cat)
builder = hm._builders[0]
self.assertEqual(builder._groups, ['0', '1', '2'])
assert_array_equal(builder._data['cat'], cat)
assert_array_equal(builder._data['cat0'], catpython)
assert_array_equal(builder._data['cat1'], catpypy)
assert_array_equal(builder._data['cat2'], catjython)
assert_array_equal(builder._data['0'], python)
assert_array_equal(builder._data['1'], pypy)
assert_array_equal(builder._data['2'], jython)
assert_array_equal(builder._data['seg_top_0'], seg_top_python)
assert_array_equal(builder._data['seg_top_1'], seg_top_pypy)
assert_array_equal(builder._data['seg_top_2'], seg_top_jython)
assert_array_equal(builder._data['z_0'], zero)
assert_array_equal(builder._data['z_1'], zero)
assert_array_equal(builder._data['z_2'], zero)
assert_array_equal(builder._data['zero'], zero)
| bsd-3-clause |
sbg2133/miscellaneous_projects | carina/ItoNH.py | 1 | 1115 | import numpy as np
import matplotlib.pyplot as plt
from astropy.io import fits
import aplpy
from astropy.wcs import WCS
import sys, os
from getIQU import IQU
from astropy import coordinates as coord
from astropy.coordinates import SkyCoord
from astropy import units as u
from scipy.interpolate import griddata
plt.ion()
root_dir = '/home/wizwit/miscellaneous_projects/carina/carinaData'
blast250_file = os.path.join(root_dir, 'smooth/3.0_arcmin/carinaneb_250_smoothed_3.0_rl.fits')
beta = 1.27
def getPsi(path_to_file):
I, Q, U, __, wcs = IQU(path_to_file)
Pvals = np.sqrt(Q**2 + U**2)
pvals = Pvals/I
# pvals /= pol_eff[band_idx]
psi = 0.5*np.arctan2(U,Q)
return I, Q, U, wcs, psi
I, __, __, wcs_250, __, = getPsi(blast250_file)
#tau_d = (nu/nu0)**beta
# See Walker pg. 71
# nu0 = frequency at which dust emission becomes optically thin
#nu0 = 0.103 * Td # 0.103 (THz/K) * Td
#Inu_dust = Bnu(Td)*(1.0 - np.exp(1.0 - e**(-1.0*tau_d))
# See Walker pg. 69
# Av = 1.086*tau_d
# N_H = 1.79e21 * Av # (atoms/cm**2 mag)
# 1) Solve tau_d for temperature
# 2) Plug into Inu_dust equation
| gpl-3.0 |
gfyoung/pandas | pandas/tests/indexes/timedeltas/test_join.py | 4 | 1497 | import numpy as np
from pandas import Index, Timedelta, timedelta_range
import pandas._testing as tm
class TestJoin:
def test_append_join_nondatetimeindex(self):
rng = timedelta_range("1 days", periods=10)
idx = Index(["a", "b", "c", "d"])
result = rng.append(idx)
assert isinstance(result[0], Timedelta)
# it works
rng.join(idx, how="outer")
def test_join_self(self, join_type):
index = timedelta_range("1 day", periods=10)
joined = index.join(index, how=join_type)
tm.assert_index_equal(index, joined)
def test_does_not_convert_mixed_integer(self):
df = tm.makeCustomDataframe(
10,
10,
data_gen_f=lambda *args, **kwargs: np.random.randn(),
r_idx_type="i",
c_idx_type="td",
)
str(df)
cols = df.columns.join(df.index, how="outer")
joined = cols.join(df.columns)
assert cols.dtype == np.dtype("O")
assert cols.dtype == joined.dtype
tm.assert_index_equal(cols, joined)
def test_join_preserves_freq(self):
# GH#32157
tdi = timedelta_range("1 day", periods=10)
result = tdi[:5].join(tdi[5:], how="outer")
assert result.freq == tdi.freq
tm.assert_index_equal(result, tdi)
result = tdi[:5].join(tdi[6:], how="outer")
assert result.freq is None
expected = tdi.delete(5)
tm.assert_index_equal(result, expected)
| bsd-3-clause |
TheChymera/LabbookDB | labbookdb/tests/test_report.py | 1 | 3726 | import pytest
from os import path
DB_PATH = '~/.demolog/meta.db'
DATA_DIR = path.join(path.dirname(path.realpath(__file__)),'../../example_data/')
def test_implant_angle_filter():
from labbookdb.report.selection import animal_id, animal_treatments, animal_operations
import numpy as np
db_path=DB_PATH
df = animal_operations(db_path)
#validate target by code
df = df[~df['OrthogonalStereotacticTarget_code'].isnull()]
df = df[df['OrthogonalStereotacticTarget_code'].str.contains('dr')]
#check pitch
df = df[~df['OrthogonalStereotacticTarget_pitch'].isin([0,np.NaN])]
animals = df['Animal_id'].tolist()
animals_eth = [animal_id(db_path,'ETH/AIC',i,reverse=True) for i in animals]
assert animals_eth == ['5684']
def test_animal_cage_treatment_control_in_report():
"""Check if animal which died before the cagetreatment was applied to its last home cage is indeed not showing a cage treatment, but still showing the animal treatment."""
from labbookdb.report.tracking import animals_info
df = animals_info(DB_PATH,
save_as=None,
functional_scan_responders=True,
treatments=True,
)
assert df[df['ETH/AIC']=='6255']['cage_treatment'].values[0] == ""
assert df[df['ETH/AIC']=='6255']['animal_treatment'].values[0] == 'aFluIV_'
def test_animal_id():
"""Check if LabbookDB animal ID is correctly reported based on external database identifier."""
from labbookdb.report.selection import animal_id
my_id = animal_id(DB_PATH,
database='ETH/AIC',
identifier='6255'
)
assert my_id == 41
def test_bids_eventsfile():
"""Check if correct BIDS events file can be sourced."""
from labbookdb.report.tracking import bids_eventsfile
import pandas as pd
df = bids_eventsfile(DB_PATH,'chr_longSOA')
bids_eventsfile = path.join(DATA_DIR,'bids_eventsfile.csv')
df_ = pd.read_csv(bids_eventsfile, index_col=0)
assert df[['onset','duration']].equals(df_[['onset','duration']])
def test_drinking_by_cage_treatment(
treatment_relative_date=True,
rounding='D',
):
from labbookdb.report.tracking import treatment_group, append_external_identifiers, qualitative_dates, cage_consumption
from labbookdb.report.selection import cage_periods, cage_drinking_measurements
known_cage_ids = [25, 38, 41]
known_consumption_values = [2.35, 2.51, 2.94, 2.95, 3.16, 3.17, 3.22, 3.23, 3.24, 3.25, 3.49, 3.63, 3.72, 4.04, 4.09, 4.58, 4.98, 5.15, 5.31, 5.39, 5.54, 5.97, 6.73, 6.78]
df = cage_drinking_measurements(DB_PATH,['cFluDW'])
df = cage_consumption(DB_PATH,df)
fuzzy_matching = {
"ofM":[-14,-15,-13,-7,-8,-6],
"ofMaF":[0,-1],
"ofMcF1":[14,13,15],
"ofMcF2":[28,27,29],
"ofMpF":[45,44,46,43,47],
}
df = qualitative_dates(df,
iterator_column='Cage_id',
date_column='relative_end_date',
label='qualitative_date',
fuzzy_matching=fuzzy_matching,
)
cage_ids = sorted(df['Cage_id'].unique())
assert cage_ids == known_cage_ids
consumption_values = df['day_animal_consumption'].values
consumption_values = [round(i, 2) for i in consumption_values]
consumption_values = sorted(list(set(consumption_values)))
assert consumption_values == known_consumption_values
def test_groups():
"""Create a `pandas.DataFrame` containing treatment and genotype group assignments"""
from labbookdb.report.tracking import treatment_group, append_external_identifiers
known_sorted_ids = [
'5667',
'5668',
'5673',
'5674',
'5675',
'5689',
'5690',
'5691',
'5692',
'5694',
'5699',
'5700',
'5704',
'5705',
'5706',
'6254',
'6256',
'6262',
]
df = treatment_group(DB_PATH, ['cFluDW','cFluDW_'], level='cage')
df = append_external_identifiers(DB_PATH, df, ['Genotype_code'])
sorted_ids = sorted(df['ETH/AIC'].tolist())
assert sorted_ids == known_sorted_ids
| bsd-3-clause |
blink1073/scikit-image | doc/examples/edges/plot_active_contours.py | 4 | 3317 | """
====================
Active Contour Model
====================
The active contour model is a method to fit open or closed splines to lines or
edges in an image. It works by minimising an energy that is in part defined by
the image and part by the spline's shape: length and smoothness. The
minimization is done implicitly in the shape energy and explicitly in the
image energy.
In the following two examples the active contour model is used (1) to segment
the face of a person from the rest of an image by fitting a closed curve
to the edges of the face and (2) to find the darkest curve between two fixed
points while obeying smoothness considerations. Typically it is a good idea to
smooth images a bit before analyzing, as done in the following examples.
.. [1] *Snakes: Active contour models*. Kass, M.; Witkin, A.; Terzopoulos, D.
International Journal of Computer Vision 1 (4): 321 (1988).
We initialize a circle around the astronaut's face and use the default boundary
condition ``bc='periodic'`` to fit a closed curve. The default parameters
``w_line=0, w_edge=1`` will make the curve search towards edges, such as the
boundaries of the face.
"""
import numpy as np
import matplotlib.pyplot as plt
from skimage.color import rgb2gray
from skimage import data
from skimage.filters import gaussian_filter
from skimage.segmentation import active_contour
# Test scipy version, since active contour is only possible
# with recent scipy version
import scipy
scipy_version = list(map(int, scipy.__version__.split('.')))
new_scipy = scipy_version[0] > 0 or \
(scipy_version[0] == 0 and scipy_version[1] >= 14)
img = data.astronaut()
img = rgb2gray(img)
s = np.linspace(0, 2*np.pi, 400)
x = 220 + 100*np.cos(s)
y = 100 + 100*np.sin(s)
init = np.array([x, y]).T
if not new_scipy:
print('You are using an old version of scipy. '
'Active contours is implemented for scipy versions '
'0.14.0 and above.')
if new_scipy:
snake = active_contour(gaussian_filter(img, 3),
init, alpha=0.015, beta=10, gamma=0.001)
fig = plt.figure(figsize=(7, 7))
ax = fig.add_subplot(111)
plt.gray()
ax.imshow(img)
ax.plot(init[:, 0], init[:, 1], '--r', lw=3)
ax.plot(snake[:, 0], snake[:, 1], '-b', lw=3)
ax.set_xticks([]), ax.set_yticks([])
ax.axis([0, img.shape[1], img.shape[0], 0])
"""
.. image:: PLOT2RST.current_figure
Here we initialize a straight line between two points, `(5, 136)` and
`(424, 50)`, and require that the spline has its end points there by giving
the boundary condition `bc='fixed'`. We furthermore make the algorithm search
for dark lines by giving a negative `w_line` value.
"""
img = data.text()
x = np.linspace(5, 424, 100)
y = np.linspace(136, 50, 100)
init = np.array([x, y]).T
if new_scipy:
snake = active_contour(gaussian_filter(img, 1), init, bc='fixed',
alpha=0.1, beta=1.0, w_line=-5, w_edge=0, gamma=0.1)
fig = plt.figure(figsize=(9, 5))
ax = fig.add_subplot(111)
plt.gray()
ax.imshow(img)
ax.plot(init[:, 0], init[:, 1], '--r', lw=3)
ax.plot(snake[:, 0], snake[:, 1], '-b', lw=3)
ax.set_xticks([]), ax.set_yticks([])
ax.axis([0, img.shape[1], img.shape[0], 0])
plt.show()
"""
.. image:: PLOT2RST.current_figure
"""
| bsd-3-clause |
KitwareMedical/ITKTubeTK | examples/archive/SegmentVesselsUsingNeuralNetworks/scripts/PreProcessing.py | 4 | 6399 | #!/usr/bin/python
###########################################################################
# PreProcessing.py :
#
# Iterate through the expert labelmap and create 65x65 patches around the
# central pixel. All positive pixels are used as positives input cases.
# The same amount of negatives is randomly picked. For each input patch,
# the corresponding filename and expected output are written to a text file
# and will be used later to create the database.
#
###########################################################################
import os
import glob
import json
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
import random
import math
# Create image set and save expected output
def createImgSet(expertImg, inputImg, filenamePrefix, fileOutputDir, textFile):
w = 32 # Patch size
count = 0 # Input count
negativeImageIndex = [] # Whole image index giving negative output
negativeIndex = [] # Vessel bound index giving negative output
# Write filename and expected output
textFile = open(textFile, "a")
textFile.truncate() # Erase file
resample = 1 # WARNING: resample pixels to reduce training size for Debug
# Iterate through the expert label map
for i in range(0, expertImg.shape[0], resample):
for j in range(0, expertImg.shape[1], resample):
if j > w and j + w + 1 < inputImg.shape[1]:
if i > w and i + w + 1 < inputImg.shape[0]:
# Centerline pixel (positive)
if expertImg[i, j] > 0.5:
count += 1
filename = os.path.join(
"1", filenamePrefix + "_" + str(i) + "_" + str(j) + ".png")
textFile.write(filename + " " + str(1) + "\n")
plt.imsave(os.path.join(fileOutputDir, filename),
inputImg[i - w:i + w + 1, j - w:j + w + 1], cmap='Greys_r')
# Vessel bound pixel (negative)
elif expertImg[i, j] > 0:
negativeIndex.append([i, j])
# Background pixel (negative)
else:
negativeImageIndex.append([i, j])
# Pick random negatives from vessel bound
rndmNegativeInd = random.sample(negativeIndex, int(math.ceil(0.8 * count)))
for [i, j] in rndmNegativeInd:
filename = os.path.join("0", filenamePrefix +
"_" + str(i) + "_" + str(j) + ".png")
textFile.write(filename + " " + str(0) + "\n")
plt.imsave(os.path.join(fileOutputDir, filename),
inputImg[i - w:i + w + 1, j - w:j + w + 1], cmap='Greys_r')
# Pick random negatives from the entire image
rndmNegativeImageInd = random.sample(
negativeImageIndex, int(math.ceil(0.2 * count)))
for [i, j] in rndmNegativeImageInd:
filename = os.path.join("0", filenamePrefix +
"_" + str(i) + "_" + str(j) + ".png")
textFile.write(filename + " " + str(0) + "\n")
plt.imsave(os.path.join(fileOutputDir, filename),
inputImg[i - w:i + w + 1, j - w:j + w + 1], cmap='Greys_r')
textFile.close()
print(count)
########
# Main #
########
# Path variable
script_params = json.load(open('params.json'))
caffe_root = script_params['CAFFE_SRC_ROOT']
hardDrive_root = script_params['CNN_DATA_ROOT']
proj_rel_path = script_params['PROJECT_REL_PATH']
caffe_proj_root = os.path.join(caffe_root, "data", proj_rel_path)
hardDrive_proj_root = os.path.join(hardDrive_root, proj_rel_path)
trainDataDir = os.path.join(hardDrive_proj_root, "training")
valDataDir = os.path.join(hardDrive_proj_root, "testing")
# Text file
trainFilename = os.path.join(caffe_proj_root, "train.txt")
trainFile = open(trainFilename, "w+")
trainFile.truncate() # Erase file
trainFile.close()
valFilename = os.path.join(caffe_proj_root, "val.txt")
valFile = open(valFilename, "w+")
valFile.truncate() # Erase file
valFile.close()
# Output patches directories
trainFileOutputDir = os.path.join(trainDataDir, "out")
if not os.path.exists(trainFileOutputDir):
os.mkdir(trainFileOutputDir)
for label in range(2):
curLabelOutputDir = os.path.join(trainFileOutputDir, str(label))
if not os.path.exists(curLabelOutputDir):
os.mkdir(curLabelOutputDir)
valFileOutputDir = os.path.join(valDataDir, "out")
if not os.path.exists(valFileOutputDir):
os.mkdir(valFileOutputDir)
for label in range(2):
curLabelOutputDir = os.path.join(valFileOutputDir, str(label))
if not os.path.exists(curLabelOutputDir):
os.mkdir(curLabelOutputDir)
# Images directories
trainExpertDir = os.path.join(trainDataDir, "expert")
trainImgDir = os.path.join(trainDataDir, "images")
valExpertDir = os.path.join(valDataDir, "expert")
valImgDir = os.path.join(valDataDir, "images")
# Create train set
trainImages = glob.glob(os.path.join(trainImgDir, "*.png"))
for trainImage in trainImages:
print(trainImage)
# Get image ID
trainImagePrefix = os.path.basename(os.path.splitext(trainImage)[0])
# Set filename
trainExpertFile = os.path.join(
trainExpertDir, trainImagePrefix + "_expert.png")
trainImageFile = os.path.join(trainImgDir, trainImagePrefix + ".png")
# Load images
trainExpert = mpimg.imread(trainExpertFile)
# print trainExpert.shape
# trainExpert=trainExpert[:,:,0]
trainImg = mpimg.imread(trainImageFile)
# trainImg=trainImg[:,:,0]
# Write images and text files
createImgSet(trainExpert, trainImg, trainImagePrefix,
trainFileOutputDir, trainFilename)
# Create validation set
valImages = glob.glob(os.path.join(valImgDir, "*.png"))
for valImage in valImages:
print(valImage)
# Get image ID
valImagePrefix = os.path.basename(os.path.splitext(valImage)[0])
# Set filename
valExpertFilename = os.path.join(
valExpertDir, valImagePrefix + "_expert.png")
valImgFilename = os.path.join(valImgDir, valImagePrefix + ".png")
# Load images
valExpert = mpimg.imread(valExpertFilename)
# valExpert=valExpert[:,:,0]
valImg = mpimg.imread(valImgFilename)
# valImg=valImg[:,:,0]
# Write images and text files
createImgSet(valExpert, valImg, valImagePrefix,
valFileOutputDir, valFilename)
| apache-2.0 |
mrshu/scikit-learn | examples/plot_permutation_test_for_classification.py | 1 | 2236 | """
=================================================================
Test with permutations the significance of a classification score
=================================================================
In order to test if a classification score is significative a technique
in repeating the classification procedure after randomizing, permuting,
the labels. The p-value is then given by the percentage of runs for
which the score obtained is greater than the classification score
obtained in the first place.
"""
# Author: Alexandre Gramfort <alexandre.gramfort@inria.fr>
# License: BSD
print __doc__
import numpy as np
import pylab as pl
from sklearn.svm import SVC
from sklearn.cross_validation import StratifiedKFold, permutation_test_score
from sklearn import datasets
from sklearn.metrics import zero_one_score
##############################################################################
# Loading a dataset
iris = datasets.load_iris()
X = iris.data
y = iris.target
n_classes = np.unique(y).size
# Some noisy data not correlated
random = np.random.RandomState(seed=0)
E = random.normal(size=(len(X), 2200))
# Add noisy data to the informative features for make the task harder
X = np.c_[X, E]
svm = SVC(kernel='linear')
cv = StratifiedKFold(y, 2)
score, permutation_scores, pvalue = permutation_test_score(
svm, X, y, zero_one_score, cv=cv, n_permutations=100, n_jobs=1)
print "Classification score %s (pvalue : %s)" % (score, pvalue)
###############################################################################
# View histogram of permutation scores
pl.hist(permutation_scores, 20, label='Permutation scores')
ylim = pl.ylim()
# BUG: vlines(..., linestyle='--') fails on older versions of matplotlib
#pl.vlines(score, ylim[0], ylim[1], linestyle='--',
# color='g', linewidth=3, label='Classification Score'
# ' (pvalue %s)' % pvalue)
#pl.vlines(1.0 / n_classes, ylim[0], ylim[1], linestyle='--',
# color='k', linewidth=3, label='Luck')
pl.plot(2 * [score], ylim, '--g', linewidth=3,
label='Classification Score'
' (pvalue %s)' % pvalue)
pl.plot(2 * [1. / n_classes], ylim, '--k', linewidth=3, label='Luck')
pl.ylim(ylim)
pl.legend()
pl.xlabel('Score')
pl.show()
| bsd-3-clause |
aruneral01/auto-sklearn | autosklearn/estimators.py | 5 | 4834 | import os
import random
import shutil
import numpy as np
import autosklearn.automl
from autosklearn.constants import *
class AutoSklearnClassifier(autosklearn.automl.AutoML):
"""This class implements the classification task. It must not be pickled!
Parameters
----------
time_left_for_this_task : int, optional (default=3600)
Time limit in seconds for the search for appropriate classification
models. By increasing this value, *auto-sklearn* will find better
configurations.
per_run_time_limit : int, optional (default=360)
Time limit for a single call to machine learning model.
initial_configurations_via_metalearning : int, optional (default=25)
ensemble_size : int, optional (default=50)
ensemble_nbest : int, optional (default=50)
seed : int, optional (default=1)
ml_memory_limit : int, optional (3000)
Memory limit for the machine learning algorithm. If the machine
learning algorithm allocates tries to allocate more memory,
its evaluation will be stopped.
"""
def __init__(self, time_left_for_this_task=3600,
per_run_time_limit=360,
initial_configurations_via_metalearning=25,
ensemble_size=50, ensemble_nbest=50, seed=1,
ml_memory_limit=3000):
random_number = random.randint(0, 10000)
pid = os.getpid()
output_dir = "/tmp/autosklearn_output_%d_%d" % (pid, random_number)
tmp_dir = "/tmp/autosklearn_tmp_%d_%d" % (pid, random_number)
os.makedirs(output_dir)
os.makedirs(tmp_dir)
super(AutoSklearnClassifier, self).__init__(
tmp_dir, output_dir, time_left_for_this_task, per_run_time_limit,
log_dir=tmp_dir,
initial_configurations_via_metalearning=initial_configurations_via_metalearning,
ensemble_size=ensemble_size, ensemble_nbest=ensemble_nbest,
seed=seed, ml_memory_limit=ml_memory_limit)
def __del__(self):
self._delete_output_directories()
def _create_output_directories(self):
os.makedirs(self.output_dir)
os.makedirs(self.tmp_dir)
def _delete_output_directories(self):
shutil.rmtree(self.tmp_dir)
shutil.rmtree(self.output_dir)
def fit(self, X, y, metric='acc_metric', feat_type=None):
"""Fit *autosklearn* to given training set (X, y).
X : array-like or sparse matrix of shape = [n_samples, n_features]
The training input samples.
y : array-like, shape = [n_samples] or [n_samples, n_outputs]
The target classes.
metric : str, optional (default='acc_metric')
The metric to optimize for. Can be one of: ['acc_metric',
'auc_metric', 'bac_metric', 'f1_metric', 'pac_metric']
feat_type : list, optional (default=None)
List of :python:`len(X.shape[1])` describing if an attribute is
continuous or categorical. Categorical attributes will
automatically 1Hot encoded.
"""
# Fit is supposed to be idempotent!
self._delete_output_directories()
self._create_output_directories()
y = np.atleast_1d(y)
if y.ndim == 1:
# reshape is necessary to preserve the data contiguity against vs
# [:, np.newaxis] that does not.
y = np.reshape(y, (-1, 1))
self.n_outputs_ = y.shape[1]
y = np.copy(y)
self.classes_ = []
self.n_classes_ = []
for k in xrange(self.n_outputs_):
classes_k, y[:, k] = np.unique(y[:, k], return_inverse=True)
self.classes_.append(classes_k)
self.n_classes_.append(classes_k.shape[0])
self.n_classes_ = np.array(self.n_classes_, dtype=np.int)
if self.n_outputs_ > 1:
task = MULTILABEL_CLASSIFICATION
else:
if len(self.classes_[0]) == 2:
task = BINARY_CLASSIFICATION
else:
task = MULTICLASS_CLASSIFICATION
# TODO: fix metafeatures calculation to allow this!
if y.shape[1] == 1:
y = y.flatten()
return super(AutoSklearnClassifier, self).fit(X, y, task, metric,
feat_type)
def predict(self, X):
"""Predict class for X.
Parameters
----------
X : array-like or sparse matrix of shape = [n_samples, n_features]
Returns
-------
y : array of shape = [n_samples] or [n_samples, n_outputs]
The predicted classes.
"""
return super(AutoSklearnClassifier, self).predict(X)
class AutoSklearnRegressor(autosklearn.automl.AutoML):
def __init__(self, **kwargs):
raise NotImplementedError() | bsd-3-clause |
aabadie/scikit-learn | examples/mixture/plot_concentration_prior.py | 25 | 5631 | """
========================================================================
Concentration Prior Type Analysis of Variation Bayesian Gaussian Mixture
========================================================================
This example plots the ellipsoids obtained from a toy dataset (mixture of three
Gaussians) fitted by the ``BayesianGaussianMixture`` class models with a
Dirichlet distribution prior
(``weight_concentration_prior_type='dirichlet_distribution'``) and a Dirichlet
process prior (``weight_concentration_prior_type='dirichlet_process'``). On
each figure, we plot the results for three different values of the weight
concentration prior.
The ``BayesianGaussianMixture`` class can adapt its number of mixture
componentsautomatically. The parameter ``weight_concentration_prior`` has a
direct link with the resulting number of components with non-zero weights.
Specifying a low value for the concentration prior will make the model put most
of the weight on few components set the remaining components weights very close
to zero. High values of the concentration prior will allow a larger number of
components to be active in the mixture.
The Dirichlet process prior allows to define an infinite number of components
and automatically selects the correct number of components: it activates a
component only if it is necessary.
On the contrary the classical finite mixture model with a Dirichlet
distribution prior will favor more uniformly weighted components and therefore
tends to divide natural clusters into unnecessary sub-components.
"""
# Author: Thierry Guillemot <thierry.guillemot.work@gmail.com>
# License: BSD 3 clause
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
from sklearn.mixture import BayesianGaussianMixture
print(__doc__)
def plot_ellipses(ax, weights, means, covars):
for n in range(means.shape[0]):
eig_vals, eig_vecs = np.linalg.eigh(covars[n])
unit_eig_vec = eig_vecs[0] / np.linalg.norm(eig_vecs[0])
angle = np.arctan2(unit_eig_vec[1], unit_eig_vec[0])
# Ellipse needs degrees
angle = 180 * angle / np.pi
# eigenvector normalization
eig_vals = 2 * np.sqrt(2) * np.sqrt(eig_vals)
ell = mpl.patches.Ellipse(means[n], eig_vals[0], eig_vals[1],
180 + angle)
ell.set_clip_box(ax.bbox)
ell.set_alpha(weights[n])
ell.set_facecolor('#56B4E9')
ax.add_artist(ell)
def plot_results(ax1, ax2, estimator, X, y, title, plot_title=False):
ax1.set_title(title)
ax1.scatter(X[:, 0], X[:, 1], s=5, marker='o', color=colors[y], alpha=0.8)
ax1.set_xlim(-2., 2.)
ax1.set_ylim(-3., 3.)
ax1.set_xticks(())
ax1.set_yticks(())
plot_ellipses(ax1, estimator.weights_, estimator.means_,
estimator.covariances_)
ax2.get_xaxis().set_tick_params(direction='out')
ax2.yaxis.grid(True, alpha=0.7)
for k, w in enumerate(estimator.weights_):
ax2.bar(k - .45, w, width=0.9, color='#56B4E9', zorder=3)
ax2.text(k, w + 0.007, "%.1f%%" % (w * 100.),
horizontalalignment='center')
ax2.set_xlim(-.6, 2 * n_components - .4)
ax2.set_ylim(0., 1.1)
ax2.tick_params(axis='y', which='both', left='off',
right='off', labelleft='off')
ax2.tick_params(axis='x', which='both', top='off')
if plot_title:
ax1.set_ylabel('Estimated Mixtures')
ax2.set_ylabel('Weight of each component')
# Parameters of the dataset
random_state, n_components, n_features = 2, 3, 2
colors = np.array(['#0072B2', '#F0E442', '#D55E00'])
covars = np.array([[[.7, .0], [.0, .1]],
[[.5, .0], [.0, .1]],
[[.5, .0], [.0, .1]]])
samples = np.array([200, 500, 200])
means = np.array([[.0, -.70],
[.0, .0],
[.0, .70]])
# mean_precision_prior= 0.8 to minimize the influence of the prior
estimators = [
("Finite mixture with a Dirichlet distribution\nprior and "
r"$\gamma_0=$", BayesianGaussianMixture(
weight_concentration_prior_type="dirichlet_distribution",
n_components=2 * n_components, reg_covar=0, init_params='random',
max_iter=1500, mean_precision_prior=.8,
random_state=random_state), [0.001, 1, 1000]),
("Infinite mixture with a Dirichlet process\n prior and" r"$\gamma_0=$",
BayesianGaussianMixture(
weight_concentration_prior_type="dirichlet_process",
n_components=2 * n_components, reg_covar=0, init_params='random',
max_iter=1500, mean_precision_prior=.8,
random_state=random_state), [1, 1000, 100000])]
# Generate data
rng = np.random.RandomState(random_state)
X = np.vstack([
rng.multivariate_normal(means[j], covars[j], samples[j])
for j in range(n_components)])
y = np.concatenate([j * np.ones(samples[j], dtype=int)
for j in range(n_components)])
# Plot results in two different figures
for (title, estimator, concentrations_prior) in estimators:
plt.figure(figsize=(4.7 * 3, 8))
plt.subplots_adjust(bottom=.04, top=0.90, hspace=.05, wspace=.05,
left=.03, right=.99)
gs = gridspec.GridSpec(3, len(concentrations_prior))
for k, concentration in enumerate(concentrations_prior):
estimator.weight_concentration_prior = concentration
estimator.fit(X)
plot_results(plt.subplot(gs[0:2, k]), plt.subplot(gs[2, k]), estimator,
X, y, r"%s$%.1e$" % (title, concentration),
plot_title=k == 0)
plt.show()
| bsd-3-clause |
bnoi/scikit-tracker | sktracker/tracker/cost_function/tests/test_abstract_cost_functions.py | 1 | 1500 | # -*- coding: utf-8 -*-
from __future__ import unicode_literals
from __future__ import division
from __future__ import absolute_import
from __future__ import print_function
from nose.tools import assert_raises
import sys
import pandas as pd
import numpy as np
from sktracker.tracker.cost_function import AbstractCostFunction
def test_abstract_cost_function():
cost_func = AbstractCostFunction(context={}, parameters={})
assert cost_func.get_block() == None
def test_abstract_cost_function_check_context():
cost_func = AbstractCostFunction(context={'cost': 1}, parameters={})
assert_raises(ValueError, cost_func.check_context, 'test_string', str)
cost_func.context['test_string'] = 5
assert_raises(TypeError, cost_func.check_context, 'test_string', str)
cost_func.context['test_string'] = "i am a string"
### This fails in py2.7
if sys.version_info[0] > 2:
cost_func.check_context('test_string', str)
assert True
def test_abstract_cost_function_check_columns():
cost_func = AbstractCostFunction(context={}, parameters={})
df = pd.DataFrame([np.arange(0, 5), np.arange(20, 25)],
columns=['x', 'y', 'z', 'w', 't'])
cost_func.check_columns(df, ['t', 'z', 'y'])
cost_func.check_columns([df], ['t', 'z', 'y'])
df = pd.DataFrame([np.arange(0, 4), np.arange(20, 24)],
columns=['x', 'y', 'w', 't'])
assert_raises(ValueError, cost_func.check_columns, df, ['t', 'z', 'y'])
| bsd-3-clause |
viisar/brew | brew/selection/dynamic/dsknn.py | 3 | 3563 | import numpy as np
from brew.base import Ensemble
from brew.metrics.diversity.paired import kuncheva_double_fault_measure
from .base import DCS
class DSKNN(DCS):
"""DS-KNN
The DS-KNN selects an ensemble of classifiers based on
their accuracy and diversity in the neighborhood of the
test sample.
Attributes
----------
`Xval` : array-like, shape = [indeterminated, n_features]
Validation set.
`yval` : array-like, shape = [indeterminated]
Labels of the validation set.
`knn` : sklearn KNeighborsClassifier,
Classifier used to find neighborhood.
Examples
--------
>>> from brew.selection.dynamic import DSKNN
>>> from brew.generation.bagging import Bagging
>>> from brew.base import EnsembleClassifier
>>>
>>> from sklearn.tree import DecisionTreeClassifier
>>> import numpy as np
>>>
>>> X = np.array([[-1, 0], [-0.8, 1], [-0.8, -1], [-0.5, 0],
[0.5, 0], [1, 0], [0.8, 1], [0.8, -1]])
>>> y = np.array([1, 1, 1, 2, 1, 2, 2, 2])
>>> tree = DecisionTreeClassifier(max_depth=1, min_samples_leaf=1)
>>> bag = Bagging(base_classifier=tree, n_classifiers=10)
>>> bag.fit(X, y)
>>>
>>> sel = DSKNN(X, y, K=3)
>>>
>>> clf = EnsembleClassifier(bag.ensemble, selector=sel)
>>> clf.predict([-1.1,-0.5])
[1]
See also
--------
brew.selection.dynamic.lca.OLA: Overall Local Accuracy.
brew.selection.dynamic.lca.LCA: Local Class Accuracy.
References
----------
Santana, Alixandre, et al. "A dynamic classifier selection method
to build ensembles using accuracy and diversity." 2006 Ninth
Brazilian Symposium on Neural Networks (SBRN'06). IEEE, 2006.
"""
def __init__(self, Xval, yval, K=5, weighted=False, knn=None,
n_1=0.7, n_2=0.3):
if n_1 < 0 or n_2 < 0 or n_1 <= n_2:
raise Exception
self.n_1 = n_1
self.n_2 = n_2
super(DSKNN, self).__init__(
Xval, yval, K=K, weighted=weighted, knn=knn)
def select(self, ensemble, x):
if ensemble.in_agreement(x):
return Ensemble([ensemble.classifiers[0]]), None
n_sel_1, n_sel_2 = self.n_1, self.n_2
if isinstance(self.n_1, float):
n_sel_1 = int(n_sel_1 * len(ensemble))
if isinstance(self.n_2, float):
n_sel_2 = int(n_sel_2 * len(ensemble))
n_sel_1 = max(n_sel_1, 1)
n_sel_2 = max(n_sel_2, 1)
# intialize variables
# the the indexes of the KNN of x
classifiers = ensemble.classifiers
[idx] = self.knn.kneighbors(x, return_distance=False)
X, y = self.Xval[idx], self.yval[idx]
acc_scores = np.array([clf.score(X, y) for clf in classifiers])
out = ensemble.output(X, mode='labels')
oracle = np.equal(out, y[:, np.newaxis])
div_scores = np.zeros(len(ensemble), dtype=float)
for i in range(len(ensemble)):
tmp = []
for j in range(len(ensemble)):
if i != j:
d = kuncheva_double_fault_measure(oracle[:, [i, j]])
tmp.append(d)
div_scores[i] = np.mean(tmp)
z = zip(np.arange(len(ensemble)), acc_scores, div_scores)
z = sorted(z, key=lambda e: e[1], reverse=True)[:n_sel_1]
z = sorted(z, key=lambda e: e[2], reverse=False)[:n_sel_2]
z = zip(*z)[0]
classifiers = [classifiers[i] for i in z]
return Ensemble(classifiers=classifiers), None
| mit |
jeffery-do/Vizdoombot | doom/lib/python3.5/site-packages/skimage/viewer/canvastools/linetool.py | 43 | 6911 | import numpy as np
from matplotlib import lines
from ...viewer.canvastools.base import CanvasToolBase, ToolHandles
__all__ = ['LineTool', 'ThickLineTool']
class LineTool(CanvasToolBase):
"""Widget for line selection in a plot.
Parameters
----------
manager : Viewer or PlotPlugin.
Skimage viewer or plot plugin object.
on_move : function
Function called whenever a control handle is moved.
This function must accept the end points of line as the only argument.
on_release : function
Function called whenever the control handle is released.
on_enter : function
Function called whenever the "enter" key is pressed.
maxdist : float
Maximum pixel distance allowed when selecting control handle.
line_props : dict
Properties for :class:`matplotlib.lines.Line2D`.
handle_props : dict
Marker properties for the handles (also see
:class:`matplotlib.lines.Line2D`).
Attributes
----------
end_points : 2D array
End points of line ((x1, y1), (x2, y2)).
"""
def __init__(self, manager, on_move=None, on_release=None, on_enter=None,
maxdist=10, line_props=None, handle_props=None,
**kwargs):
super(LineTool, self).__init__(manager, on_move=on_move,
on_enter=on_enter,
on_release=on_release, **kwargs)
props = dict(color='r', linewidth=1, alpha=0.4, solid_capstyle='butt')
props.update(line_props if line_props is not None else {})
self.linewidth = props['linewidth']
self.maxdist = maxdist
self._active_pt = None
x = (0, 0)
y = (0, 0)
self._end_pts = np.transpose([x, y])
self._line = lines.Line2D(x, y, visible=False, animated=True, **props)
self.ax.add_line(self._line)
self._handles = ToolHandles(self.ax, x, y,
marker_props=handle_props)
self._handles.set_visible(False)
self.artists = [self._line, self._handles.artist]
if on_enter is None:
def on_enter(pts):
x, y = np.transpose(pts)
print("length = %0.2f" %
np.sqrt(np.diff(x)**2 + np.diff(y)**2))
self.callback_on_enter = on_enter
self.manager.add_tool(self)
@property
def end_points(self):
return self._end_pts.astype(int)
@end_points.setter
def end_points(self, pts):
self._end_pts = np.asarray(pts)
self._line.set_data(np.transpose(pts))
self._handles.set_data(np.transpose(pts))
self._line.set_linewidth(self.linewidth)
self.set_visible(True)
self.redraw()
def hit_test(self, event):
if event.button != 1 or not self.ax.in_axes(event):
return False
idx, px_dist = self._handles.closest(event.x, event.y)
if px_dist < self.maxdist:
self._active_pt = idx
return True
else:
self._active_pt = None
return False
def on_mouse_press(self, event):
self.set_visible(True)
if self._active_pt is None:
self._active_pt = 0
x, y = event.xdata, event.ydata
self._end_pts = np.array([[x, y], [x, y]])
def on_mouse_release(self, event):
if event.button != 1:
return
self._active_pt = None
self.callback_on_release(self.geometry)
self.redraw()
def on_move(self, event):
if event.button != 1 or self._active_pt is None:
return
if not self.ax.in_axes(event):
return
self.update(event.xdata, event.ydata)
self.callback_on_move(self.geometry)
def update(self, x=None, y=None):
if x is not None:
self._end_pts[self._active_pt, :] = x, y
self.end_points = self._end_pts
@property
def geometry(self):
return self.end_points
class ThickLineTool(LineTool):
"""Widget for line selection in a plot.
The thickness of the line can be varied using the mouse scroll wheel, or
with the '+' and '-' keys.
Parameters
----------
manager : Viewer or PlotPlugin.
Skimage viewer or plot plugin object.
on_move : function
Function called whenever a control handle is moved.
This function must accept the end points of line as the only argument.
on_release : function
Function called whenever the control handle is released.
on_enter : function
Function called whenever the "enter" key is pressed.
on_change : function
Function called whenever the line thickness is changed.
maxdist : float
Maximum pixel distance allowed when selecting control handle.
line_props : dict
Properties for :class:`matplotlib.lines.Line2D`.
handle_props : dict
Marker properties for the handles (also see
:class:`matplotlib.lines.Line2D`).
Attributes
----------
end_points : 2D array
End points of line ((x1, y1), (x2, y2)).
"""
def __init__(self, manager, on_move=None, on_enter=None, on_release=None,
on_change=None, maxdist=10, line_props=None, handle_props=None):
super(ThickLineTool, self).__init__(manager,
on_move=on_move,
on_enter=on_enter,
on_release=on_release,
maxdist=maxdist,
line_props=line_props,
handle_props=handle_props)
if on_change is None:
def on_change(*args):
pass
self.callback_on_change = on_change
def on_scroll(self, event):
if not event.inaxes:
return
if event.button == 'up':
self._thicken_scan_line()
elif event.button == 'down':
self._shrink_scan_line()
def on_key_press(self, event):
if event.key == '+':
self._thicken_scan_line()
elif event.key == '-':
self._shrink_scan_line()
def _thicken_scan_line(self):
self.linewidth += 1
self.update()
self.callback_on_change(self.geometry)
def _shrink_scan_line(self):
if self.linewidth > 1:
self.linewidth -= 1
self.update()
self.callback_on_change(self.geometry)
if __name__ == '__main__': # pragma: no cover
from ... import data
from ...viewer import ImageViewer
image = data.camera()
viewer = ImageViewer(image)
h, w = image.shape
line_tool = ThickLineTool(viewer)
line_tool.end_points = ([w/3, h/2], [2*w/3, h/2])
viewer.show()
| mit |
CopyChat/Plotting | Python/PythonNetCDF.py | 1 | 10821 | '''
NAME
NetCDF with Python
PURPOSE
To demonstrate how to read and write data with NetCDF files using
a NetCDF file from the NCEP/NCAR Reanalysis.
Plotting using Matplotlib and Basemap is also shown.
PROGRAMMER(S)
Chris Slocum
REVISION HISTORY
20140320 -- Initial version created and posted online
20140722 -- Added basic error handling to ncdump
Thanks to K.-Michael Aye for highlighting the issue
REFERENCES
netcdf4-python -- http://code.google.com/p/netcdf4-python/
NCEP/NCAR Reanalysis -- Kalnay et al. 1996
http://dx.doi.org/10.1175/1520-0477(1996)077<0437:TNYRP>2.0.CO;2
'''
import datetime as dt # Python standard library datetime module
import numpy as np
from netCDF4 import Dataset # http://code.google.com/p/netcdf4-python/
import matplotlib.pyplot as plt
from mpl_toolkits.basemap import Basemap, addcyclic, shiftgrid
def ncdump(nc_fid, verb=True):
'''
ncdump outputs dimensions, variables and their attribute information.
The information is similar to that of NCAR's ncdump utility.
ncdump requires a valid instance of Dataset.
Parameters
----------
nc_fid : netCDF4.Dataset
A netCDF4 dateset object
verb : Boolean
whether or not nc_attrs, nc_dims, and nc_vars are printed
Returns
-------
nc_attrs : list
A Python list of the NetCDF file global attributes
nc_dims : list
A Python list of the NetCDF file dimensions
nc_vars : list
A Python list of the NetCDF file variables
'''
def print_ncattr(key):
"""
Prints the NetCDF file attributes for a given key
Parameters
----------
key : unicode
a valid netCDF4.Dataset.variables key
"""
try:
print "\t\ttype:", repr(nc_fid.variables[key].dtype)
for ncattr in nc_fid.variables[key].ncattrs():
print '\t\t%s:' % ncattr,\
repr(nc_fid.variables[key].getncattr(ncattr))
except KeyError:
print "\t\tWARNING: %s does not contain variable attributes" % key
# NetCDF global attributes
nc_attrs = nc_fid.ncattrs()
if verb:
print "NetCDF Global Attributes:"
for nc_attr in nc_attrs:
print '\t%s:' % nc_attr, repr(nc_fid.getncattr(nc_attr))
nc_dims = [dim for dim in nc_fid.dimensions] # list of nc dimensions
# Dimension shape information.
if verb:
print "NetCDF dimension information:"
for dim in nc_dims:
print "\tName:", dim
print "\t\tsize:", len(nc_fid.dimensions[dim])
print_ncattr(dim)
# Variable information.
nc_vars = [var for var in nc_fid.variables] # list of nc variables
if verb:
print "NetCDF variable information:"
for var in nc_vars:
if var not in nc_dims:
print '\tName:', var
print "\t\tdimensions:", nc_fid.variables[var].dimensions
print "\t\tsize:", nc_fid.variables[var].size
print_ncattr(var)
return nc_attrs, nc_dims, nc_vars
nc_f = './CLM45_Micro_UW_SRF.2005120100.for.test.nc' # Your filename
nc_fid = Dataset(nc_f, 'r') # Dataset is the class behavior to open the file
# and create an instance of the ncCDF4 class
nc_attrs, nc_dims, nc_vars = ncdump(nc_fid)
# Extract data from NetCDF file
lats = nc_fid.variables['xlat'][:] # extract/copy the data
lons = nc_fid.variables['xlon'][:]
time = nc_fid.variables['time'][:]
rsds = nc_fid.variables['rsds'][:] # shape is time, lat, lon as shown above
time_idx = 237 # some random day in 2012
# Python and the renalaysis are slightly off in time so this fixes that problem
offset = dt.timedelta(hours=48)
# List of all times in the file as datetime objects
dt_time = [dt.date(1, 1, 1) + dt.timedelta(hours=t/20) - offset\
for t in time]
cur_time = dt_time[time_idx]
# Plot of global temperature on our random day
fig = plt.figure()
fig.subplots_adjust(left=0., right=1., bottom=0., top=0.9)
# Setup the map. See http://matplotlib.org/basemap/users/mapsetup.html
# for other projections.
m = Basemap(projection='moll', llcrnrlat=-90, urcrnrlat=90,\
llcrnrlon=0, urcrnrlon=360, resolution='c', lon_0=0)
m.drawcoastlines()
m.drawmapboundary()
# Make the plot continuous
test=rsds[0,:,:]
print test.shape
print rsds.shape
print lons.shape
rsds_cyclic, lons_cyclic = addcyclic(rsds[time_idx,:,:], lons)
# Shift the grid so lons go from -180 to 180 instead of 0 to 360.
rsds_cyclic, lons_cyclic = shiftgrid(180., rsds_cyclic, lons_cyclic, start=False)
# Create 2D lat/lon arrays for Basemap
lon2d, lat2d = np.meshgrid(lons_cyclic, lats)
# Transforms lat/lon into plotting coordinates for projection
x, y = m(lon2d, lat2d)
# Plot of rsds temperature with 11 contour intervals
cs = m.contourf(x, y, rsds_cyclic, 11, cmap=plt.cm.Spectral_r)
cbar = plt.colorbar(cs, orientation='horizontal', shrink=0.5)
cbar.set_label("%s (%s)" % (nc_fid.variables['rsds'].var_desc,\
nc_fid.variables['rsds'].units))
plt.title("%s on %s" % (nc_fid.variables['rsds'].var_desc, cur_time))
# Writing NetCDF files
# For this example, we will create two NetCDF4 files. One with the global rsds
# temperature departure from its value at Darwin, Australia. The other with
# the temperature profile for the entire year at Darwin.
darwin = {'name': 'Darwin, Australia', 'lat': -12.45, 'lon': 130.83}
# Find the nearest latitude and longitude for Darwin
lat_idx = np.abs(lats - darwin['lat']).argmin()
lon_idx = np.abs(lons - darwin['lon']).argmin()
# Simple example: temperature profile for the entire year at Darwin.
# Open a new NetCDF file to write the data to. For format, you can choose from
# 'NETCDF3_CLASSIC', 'NETCDF3_64BIT', 'NETCDF4_CLASSIC', and 'NETCDF4'
w_nc_fid = Dataset('darwin_2012.nc', 'w', format='NETCDF4')
w_nc_fid.description = "NCEP/NCAR Reanalysis %s from its value at %s. %s" %\
(nc_fid.variables['rsds'].var_desc.lower(),\
darwin['name'], nc_fid.description)
# Using our previous dimension info, we can create the new time dimension
# Even though we know the size, we are going to set the size to unknown
w_nc_fid.createDimension('time', None)
w_nc_dim = w_nc_fid.createVariable('time', nc_fid.variables['time'].dtype,\
('time',))
# You can do this step yourself but someone else did the work for us.
for ncattr in nc_fid.variables['time'].ncattrs():
w_nc_dim.setncattr(ncattr, nc_fid.variables['time'].getncattr(ncattr))
# Assign the dimension data to the new NetCDF file.
w_nc_fid.variables['time'][:] = time
w_nc_var = w_nc_fid.createVariable('rsds', 'f8', ('time'))
w_nc_var.setncatts({'long_name': u"mean Daily Air temperature",\
'units': u"degK", 'level_desc': u'Surface',\
'var_desc': u"Air temperature",\
'statistic': u'Mean\nM'})
w_nc_fid.variables['rsds'][:] = rsds[time_idx, lat_idx, lon_idx]
w_nc_fid.close() # close the new file
# A plot of the temperature profile for Darwin in 2012
fig = plt.figure()
plt.plot(dt_time, rsds[:, lat_idx, lon_idx], c='r')
plt.plot(dt_time[time_idx], rsds[time_idx, lat_idx, lon_idx], c='b', marker='o')
plt.text(dt_time[time_idx], rsds[time_idx, lat_idx, lon_idx], cur_time,\
ha='right')
fig.autofmt_xdate()
plt.ylabel("%s (%s)" % (nc_fid.variables['rsds'].var_desc,\
nc_fid.variables['rsds'].units))
plt.xlabel("Time")
plt.title("%s from\n%s for %s" % (nc_fid.variables['rsds'].var_desc,\
darwin['name'], cur_time.year))
# Complex example: global temperature departure from its value at Darwin
departure = rsds[:, :, :] - rsds[:, lat_idx, lon_idx].reshape((time.shape[0],\
1, 1))
# Open a new NetCDF file to write the data to. For format, you can choose from
# 'NETCDF3_CLASSIC', 'NETCDF3_64BIT', 'NETCDF4_CLASSIC', and 'NETCDF4'
w_nc_fid = Dataset('rsds.departure.sig995.2012.nc', 'w', format='NETCDF4')
w_nc_fid.description = "The departure of the NCEP/NCAR Reanalysis " +\
"%s from its value at %s. %s" %\
(nc_fid.variables['rsds'].var_desc.lower(),\
darwin['name'], nc_fid.description)
# Using our previous dimension information, we can create the new dimensions
data = {}
for dim in nc_dims:
w_nc_fid.createDimension(dim, nc_fid.variables[dim].size)
data[dim] = w_nc_fid.createVariable(dim, nc_fid.variables[dim].dtype,\
(dim,))
# You can do this step yourself but someone else did the work for us.
for ncattr in nc_fid.variables[dim].ncattrs():
data[dim].setncattr(ncattr, nc_fid.variables[dim].getncattr(ncattr))
# Assign the dimension data to the new NetCDF file.
w_nc_fid.variables['time'][:] = time
w_nc_fid.variables['lat'][:] = lats
w_nc_fid.variables['lon'][:] = lons
# Ok, time to create our departure variable
w_nc_var = w_nc_fid.createVariable('rsds_dep', 'f8', ('time', 'lat', 'lon'))
w_nc_var.setncatts({'long_name': u"mean Daily Air temperature departure",\
'units': u"degK", 'level_desc': u'Surface',\
'var_desc': u"Air temperature departure",\
'statistic': u'Mean\nM'})
w_nc_fid.variables['rsds_dep'][:] = departure
w_nc_fid.close() # close the new file
# Rounded maximum absolute value of the departure used for contouring
max_dep = np.round(np.abs(departure[time_idx, :, :]).max()+5., decimals=-1)
# Generate a figure of the departure for a single day
fig = plt.figure()
fig.subplots_adjust(left=0., right=1., bottom=0., top=0.9)
m = Basemap(projection='moll', llcrnrlat=-90, urcrnrlat=90,\
llcrnrlon=0, urcrnrlon=360, resolution='c', lon_0=0)
m.drawcoastlines()
m.drawmapboundary()
dep_cyclic, lons_cyclic = addcyclic(departure[time_idx, :, :], lons)
dep_cyclic, lons_cyclic = shiftgrid(180., dep_cyclic, lons_cyclic, start=False)
lon2d, lat2d = np.meshgrid(lons_cyclic, lats)
x, y = m(lon2d, lat2d)
levels = np.linspace(-max_dep, max_dep, 11)
cs = m.contourf(x, y, dep_cyclic, levels=levels, cmap=plt.cm.bwr)
x, y = m(darwin['lon'], darwin['lat'])
plt.plot(x, y, c='c', marker='o')
plt.text(x, y, 'Darwin,\nAustralia', color='r', weight='semibold')
cbar = plt.colorbar(cs, orientation='horizontal', shrink=0.5)
cbar.set_label("%s departure (%s)" % (nc_fid.variables['rsds'].var_desc,\
nc_fid.variables['rsds'].units))
plt.title("Departure of Global %s from\n%s for %s" %\
(nc_fid.variables['rsds'].var_desc, darwin['name'], cur_time))
plt.show()
# Close original NetCDF file.
nc_fid.close()
| gpl-3.0 |
ephes/scikit-learn | sklearn/covariance/__init__.py | 389 | 1157 | """
The :mod:`sklearn.covariance` module includes methods and algorithms to
robustly estimate the covariance of features given a set of points. The
precision matrix defined as the inverse of the covariance is also estimated.
Covariance estimation is closely related to the theory of Gaussian Graphical
Models.
"""
from .empirical_covariance_ import empirical_covariance, EmpiricalCovariance, \
log_likelihood
from .shrunk_covariance_ import shrunk_covariance, ShrunkCovariance, \
ledoit_wolf, ledoit_wolf_shrinkage, \
LedoitWolf, oas, OAS
from .robust_covariance import fast_mcd, MinCovDet
from .graph_lasso_ import graph_lasso, GraphLasso, GraphLassoCV
from .outlier_detection import EllipticEnvelope
__all__ = ['EllipticEnvelope',
'EmpiricalCovariance',
'GraphLasso',
'GraphLassoCV',
'LedoitWolf',
'MinCovDet',
'OAS',
'ShrunkCovariance',
'empirical_covariance',
'fast_mcd',
'graph_lasso',
'ledoit_wolf',
'ledoit_wolf_shrinkage',
'log_likelihood',
'oas',
'shrunk_covariance']
| bsd-3-clause |
equialgo/scikit-learn | examples/hetero_feature_union.py | 81 | 6241 | """
=============================================
Feature Union with Heterogeneous Data Sources
=============================================
Datasets can often contain components of that require different feature
extraction and processing pipelines. This scenario might occur when:
1. Your dataset consists of heterogeneous data types (e.g. raster images and
text captions)
2. Your dataset is stored in a Pandas DataFrame and different columns
require different processing pipelines.
This example demonstrates how to use
:class:`sklearn.feature_extraction.FeatureUnion` on a dataset containing
different types of features. We use the 20-newsgroups dataset and compute
standard bag-of-words features for the subject line and body in separate
pipelines as well as ad hoc features on the body. We combine them (with
weights) using a FeatureUnion and finally train a classifier on the combined
set of features.
The choice of features is not particularly helpful, but serves to illustrate
the technique.
"""
# Author: Matt Terry <matt.terry@gmail.com>
#
# License: BSD 3 clause
from __future__ import print_function
import numpy as np
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.datasets import fetch_20newsgroups
from sklearn.datasets.twenty_newsgroups import strip_newsgroup_footer
from sklearn.datasets.twenty_newsgroups import strip_newsgroup_quoting
from sklearn.decomposition import TruncatedSVD
from sklearn.feature_extraction import DictVectorizer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics import classification_report
from sklearn.pipeline import FeatureUnion
from sklearn.pipeline import Pipeline
from sklearn.svm import SVC
class ItemSelector(BaseEstimator, TransformerMixin):
"""For data grouped by feature, select subset of data at a provided key.
The data is expected to be stored in a 2D data structure, where the first
index is over features and the second is over samples. i.e.
>> len(data[key]) == n_samples
Please note that this is the opposite convention to scikit-learn feature
matrixes (where the first index corresponds to sample).
ItemSelector only requires that the collection implement getitem
(data[key]). Examples include: a dict of lists, 2D numpy array, Pandas
DataFrame, numpy record array, etc.
>> data = {'a': [1, 5, 2, 5, 2, 8],
'b': [9, 4, 1, 4, 1, 3]}
>> ds = ItemSelector(key='a')
>> data['a'] == ds.transform(data)
ItemSelector is not designed to handle data grouped by sample. (e.g. a
list of dicts). If your data is structured this way, consider a
transformer along the lines of `sklearn.feature_extraction.DictVectorizer`.
Parameters
----------
key : hashable, required
The key corresponding to the desired value in a mappable.
"""
def __init__(self, key):
self.key = key
def fit(self, x, y=None):
return self
def transform(self, data_dict):
return data_dict[self.key]
class TextStats(BaseEstimator, TransformerMixin):
"""Extract features from each document for DictVectorizer"""
def fit(self, x, y=None):
return self
def transform(self, posts):
return [{'length': len(text),
'num_sentences': text.count('.')}
for text in posts]
class SubjectBodyExtractor(BaseEstimator, TransformerMixin):
"""Extract the subject & body from a usenet post in a single pass.
Takes a sequence of strings and produces a dict of sequences. Keys are
`subject` and `body`.
"""
def fit(self, x, y=None):
return self
def transform(self, posts):
features = np.recarray(shape=(len(posts),),
dtype=[('subject', object), ('body', object)])
for i, text in enumerate(posts):
headers, _, bod = text.partition('\n\n')
bod = strip_newsgroup_footer(bod)
bod = strip_newsgroup_quoting(bod)
features['body'][i] = bod
prefix = 'Subject:'
sub = ''
for line in headers.split('\n'):
if line.startswith(prefix):
sub = line[len(prefix):]
break
features['subject'][i] = sub
return features
pipeline = Pipeline([
# Extract the subject & body
('subjectbody', SubjectBodyExtractor()),
# Use FeatureUnion to combine the features from subject and body
('union', FeatureUnion(
transformer_list=[
# Pipeline for pulling features from the post's subject line
('subject', Pipeline([
('selector', ItemSelector(key='subject')),
('tfidf', TfidfVectorizer(min_df=50)),
])),
# Pipeline for standard bag-of-words model for body
('body_bow', Pipeline([
('selector', ItemSelector(key='body')),
('tfidf', TfidfVectorizer()),
('best', TruncatedSVD(n_components=50)),
])),
# Pipeline for pulling ad hoc features from post's body
('body_stats', Pipeline([
('selector', ItemSelector(key='body')),
('stats', TextStats()), # returns a list of dicts
('vect', DictVectorizer()), # list of dicts -> feature matrix
])),
],
# weight components in FeatureUnion
transformer_weights={
'subject': 0.8,
'body_bow': 0.5,
'body_stats': 1.0,
},
)),
# Use a SVC classifier on the combined features
('svc', SVC(kernel='linear')),
])
# limit the list of categories to make running this example faster.
categories = ['alt.atheism', 'talk.religion.misc']
train = fetch_20newsgroups(random_state=1,
subset='train',
categories=categories,
)
test = fetch_20newsgroups(random_state=1,
subset='test',
categories=categories,
)
pipeline.fit(train.data, train.target)
y = pipeline.predict(test.data)
print(classification_report(y, test.target))
| bsd-3-clause |
mirestrepo/voxels-at-lems | registration_eval/results/compute_trans_geo_accuracy.py | 1 | 13935 | #!/usr/bin/env python
# encoding: utf-8
"""
compute_transformation_error.py
Created by Maria Isabel Restrepo on 2012-09-24.
Copyright (c) 2012 . All rights reserved.
This script computes the distances betweeen an estimated similarity transformation and its ground truth
The transformation is used to transform a "source" coordinate system into a "target coordinate system"
To compute the error between the translations, the L2 norm diference translation vectors in the
"source coordinate system" is computed. Since distances are preserved under R and T, only scale is applied.
The rotation error is computed as the half angle between the normalized queternions i.e acos(|<q1,q2>|) in [0, pi/2]
"""
import os
import sys
import logging
import argparse
import vpcl_adaptor as vpcl
import numpy as np
from numpy import linalg as LA
import transformations as tf
import math
import matplotlib.pyplot as plt
sys.path.append(os.pardir)
import reg3d_transformations as reg3d_T
LOG = None
"""Compute the accuracy between the LIDAR fiducial points
and corresponding geo-register correspondances"""
def compute_ref_accuracy(fid_path, original_corrs_path,
geo_tform):
#Load fiducial .ply
fid = open(fid_path, 'r')
fid_points = np.genfromtxt(fid, dtype=float, delimiter=' ',
skip_header=9)
fid.close()
#Load original corrs .ply
fid = open(original_corrs_path, 'r')
original_corrs = np.genfromtxt(fid, dtype=float,
delimiter=' ', skip_header=9)
fid.close()
#Load transformation
#************GEO**************"
Tfis = open(geo_tform, 'r')
lines = []
lines = Tfis.readlines()
scale_geo = float(lines[0])
Ss_geo = tf.scale_matrix(scale_geo)
quat_line = lines[1].split(" ")
quat_geo = np.array([float(quat_line[3]), float(quat_line[0]),
float(quat_line[1]), float(quat_line[2])])
Rs_geo = tf.quaternion_matrix(quat_geo)
trans_line = lines[2].split(" ")
trans_geo = np.array([float(trans_line[0]), float(trans_line[1]),
float(trans_line[2])])
Tfis.close()
Hs_geo = Rs_geo.copy()
Hs_geo[:3, 3] = trans_geo[:3]
Hs_geo = Ss_geo.dot(Hs_geo)
LOG.debug("\n******Geo***** \n Scale: \n%s \nR:\n%s \nT:\n%s \nH:\n%s",
Ss_geo, Rs_geo, trans_geo, Hs_geo)
#Compute the "reference error"
#i.e. fiducial points - geo registered correspondances
npoints, c = fid_points.shape
if npoints != 30:
LOG.warn("Number of fiducial point is NOT 30")
if c != 3:
LOG.error("Fiducial points has the wrong number of dimensions")
# import code; code.interact(local=locals())
fid_points_hom = np.hstack((fid_points, np.ones([npoints, 1]))).T
original_corrs_hom = np.hstack((original_corrs, np.ones([npoints, 1]))).T
geo_corrs_hom = Hs_geo.dot(original_corrs_hom)
geo_ref_diff = geo_corrs_hom - fid_points_hom
# import pdb; pdb.set_trace()
delta_z = np.sqrt(geo_ref_diff[2, :] * geo_ref_diff[2, :])
delta_r = np.sqrt(geo_ref_diff[0, :] * geo_ref_diff[0, :] +
geo_ref_diff[1, :] * geo_ref_diff[1, :])
return delta_z, delta_r
def compute_geo_accuracy(fid_path, original_corrs_path,
geo_tform, trials_root, desc_name,
niter, ntrials, percentile=99):
#Load fiducial .ply
fid = open(fid_path, 'r')
fid_points = np.genfromtxt(fid, delimiter=' ',
skip_header=9)
fid.close()
#Load original corrs .ply
fid = open(original_corrs_path, 'r')
original_corrs = np.genfromtxt(fid, delimiter=' ', skip_header=9)
fid.close()
#load the geo tranformation
GEO = reg3d_T.geo_transformation(geo_tform);
#Compute the "reference error"
#i.e. fiducial points - geo registered correspondances
npoints, c = fid_points.shape
if npoints != 30:
LOG.warn("Number of fiducial point is NOT 30")
if c != 3:
LOG.error("Fiducial points has the wrong number of dimensions")
# import code; code.interact(local=locals())
fid_points_hom = np.hstack((fid_points, np.ones([npoints, 1]))).T
original_corrs_hom = np.hstack((original_corrs, np.ones([npoints, 1]))).T
geo_corrs_hom = GEO.transform_points(original_corrs_hom)
geo_ref_diff = geo_corrs_hom - fid_points_hom
# import pdb; pdb.set_trace()
delta_z = (geo_ref_diff[2, :] **2) ** (1./2.)
delta_r = (geo_ref_diff[0, :] **2 + geo_ref_diff[1, :] **2 )** (1./2.)
delta_z_ia = np.zeros([ntrials, npoints])
delta_r_ia = np.zeros([ntrials, npoints])
delta_z_icp = np.zeros([ntrials, npoints])
delta_r_icp = np.zeros([ntrials, npoints])
for trial in range(0, ntrials):
print "********Trial", trial, "**********"
#Load the transformations for this trial
#************Hs**************#
#read source to target "Ground Truth" Transformation
Tfile = trials_root + "/trial_" + str(trial) + "/Hs_inv.txt"
GT_Tform = reg3d_T.gt_transformation(Tfile)
src_features_dir = (trials_root + "/trial_" + str(trial) +
"/" + desc_name)
Tfile_ia = (src_features_dir + "/ia_transformation_" +
str(percentile) + "_" + str(niter) + ".txt")
Tfile_icp = (src_features_dir + "/icp_transformation_" +
str(percentile) + "_" + str(niter) + ".txt")
REG_Tform = reg3d_T.pcl_transformation(Tfile_ia, Tfile_icp)
Hs_ia_error = REG_Tform.Hs_ia.dot(GT_Tform.Hs)
Hs_icp_error = REG_Tform.Hs_icp.dot(GT_Tform.Hs)
# transform the points with the residual transformations
ia_corrs_hom = Hs_ia_error.dot(original_corrs_hom)
icp_corrs_hom = Hs_icp_error.dot(original_corrs_hom)
# geo-register
geo_ia_corrs_hom = GEO.transform_points(ia_corrs_hom)
geo_icp_corrs_hom = GEO.transform_points(icp_corrs_hom)
# distances
geo_ia_ref_diff = geo_ia_corrs_hom - fid_points_hom
geo_icp_ref_diff = geo_icp_corrs_hom - fid_points_hom
delta_z_ia[trial, :] = np.sqrt(geo_ia_ref_diff[2, :] ** 2)
delta_r_ia[trial, :] = np.sqrt(geo_ia_ref_diff[0, :] ** 2 +
geo_ia_ref_diff[1, :] ** 2 )
delta_z_icp[trial, :] = np.sqrt(geo_icp_ref_diff[2, :] ** 2)
delta_r_icp[trial, :] = np.sqrt(geo_icp_ref_diff[0, :] ** 2 +
geo_icp_ref_diff[1, :] ** 2)
# import pdb; pdb.set_trace()
return delta_z, delta_r,\
delta_z_ia, delta_r_ia, \
delta_z_icp, delta_r_icp
def main(logfile=None):
global LOG
LOG = setlogging(logfile)
descriptors = ["FPFH_30", "SHOT_30"]
niter = 500;
ntrials = 10;
plot_errors = True;
if (plot_errors):
colors = ['magenta','green'];
markers = ['o', 's', '*', '+', '^', 'v']
fid_path = "/data/lidar_providence/downtown_offset-1-financial-dan-pts1.ply"
original_corrs_path = "/data/lidar_providence/downtown_offset-1-financial-dan-pts0.ply"
trials_root = "/Users/isa/Experiments/reg3d_eval/downtown_dan";
geo_tform = "/data/lidar_providence/downtown_offset-1-financial-dan-Hs.txt"
for d_idx in range(0, len(descriptors)):
desc_name = descriptors[d_idx]
delta_z, delta_r, \
delta_z_ia, delta_r_ia, \
delta_z_icp, delta_r_icp = compute_geo_accuracy(fid_path,
original_corrs_path,
geo_tform, trials_root, desc_name,
niter, ntrials)
#sort errors for all trials to get the 70 80 90 % errors
delta_z_ia.sort(axis=0)
delta_r_ia.sort(axis=0)
delta_z_icp.sort(axis=0)
delta_r_icp.sort(axis=0)
CE_70_ia = delta_r_ia[int(0.7 * ntrials) - 1, :]
CE_80_ia = delta_r_ia[int(0.8 * ntrials) - 1, :]
CE_90_ia = delta_r_ia[int(0.9 * ntrials) - 1, :]
LE_70_ia = delta_z_ia[int(0.7 * ntrials) - 1, :]
LE_80_ia = delta_z_ia[int(0.8 * ntrials) - 1, :]
LE_90_ia = delta_z_ia[int(0.9 * ntrials) - 1, :]
CE_70_icp = delta_r_icp[int(0.7 * ntrials) - 1, :]
CE_80_icp = delta_r_icp[int(0.8 * ntrials) - 1, :]
CE_90_icp = delta_r_icp[int(0.9 * ntrials) - 1, :]
LE_70_icp = delta_z_icp[int(0.7 * ntrials) - 1, :]
LE_80_icp = delta_z_icp[int(0.8 * ntrials) - 1, :]
LE_90_icp = delta_z_icp[int(0.9 * ntrials) - 1, :]
if (plot_errors):
#Plot CE and LE
fig_ia_CE = plt.figure()
ax_ia_CE = fig_ia_CE.add_subplot(111);
plt.hold(True);
plt.axis(tight=True);
ax_ia_CE.plot(CE_70_ia, "--s", color="green", label= "CE_70");
ax_ia_CE.plot(CE_80_ia, "--^", color="magenta", label= "CE_80");
ax_ia_CE.plot(CE_90_ia, "--*", color="blue", label= "CE_90");
ax_ia_CE.plot( delta_r, "--o", color="cyan", label= "GT");
ax_ia_CE.set_xlabel('Fiducial Marker (index)',fontsize= 20);
ax_ia_CE.set_ylabel('Error (meters)',fontsize= 20);
ax_ia_CE.legend(loc='best', frameon=False);
# ax_ia_CE.set_title('IA CE')
fname = trials_root + "/GEO_results/IA_CE_" + desc_name + ".pdf"
fig_ia_CE.savefig(fname, transparent=True, pad_inches=5)
fig_ia_LE = plt.figure()
ax_ia_LE = fig_ia_LE.add_subplot(111);
plt.hold(True);
plt.axis(tight=True);
ax_ia_LE.plot(LE_70_ia, "--s", color="green", label= "LE_70");
ax_ia_LE.plot(LE_80_ia, "--^", color="magenta", label= "LE_80");
ax_ia_LE.plot(LE_90_ia, "--*", color="blue", label= "LE_90");
ax_ia_LE.plot( delta_z, "--o", color="cyan", label= "GT");
ax_ia_LE.set_xlabel('Fiducial Marker (index)',fontsize= 20);
ax_ia_LE.set_ylabel('Error (meters)',fontsize= 20);
ax_ia_LE.legend(loc='best', frameon=False);
# ax_ia_LE.set_title('IA LE')
fname = trials_root + "/GEO_results/IA_LE_" + desc_name + ".pdf"
fig_ia_LE.savefig(fname, transparent=True, pad_inches=5)
fig_icp_CE = plt.figure()
ax_icp_CE = fig_icp_CE.add_subplot(111);
plt.hold(True);
plt.axis(tight=True);
ax_icp_CE.plot(CE_70_icp, "--s", color="green", label= "CE_70");
ax_icp_CE.plot(CE_80_icp, "--^", color="magenta", label= "CE_80");
ax_icp_CE.plot(CE_90_icp, "--*", color="blue", label= "CE_90");
ax_icp_CE.plot( delta_r, "--o", color="cyan", label= "GT");
ax_icp_CE.set_xlabel('Fiducial Marker (index)',fontsize= 20);
ax_icp_CE.set_ylabel('Error (meters)',fontsize= 20);
ax_icp_CE.legend(loc='best', frameon=False);
# ax_icp_CE.set_title('ICP CE')
fname = trials_root + "/GEO_results/ICP_CE_" + desc_name + ".pdf"
fig_icp_CE.savefig(fname, transparent=True, pad_inches=5)
fig_icp_LE = plt.figure()
ax_icp_LE = fig_icp_LE.add_subplot(111);
plt.hold(True);
plt.axis(tight=True);
ax_icp_LE.plot(LE_70_icp, "--s", color="green", label= "LE_70");
ax_icp_LE.plot(LE_80_icp, "--^", color="magenta", label= "LE_80");
ax_icp_LE.plot(LE_90_icp, "--*", color="blue", label= "LE_90");
ax_icp_LE.plot( delta_z, "--o", color="cyan", label= "GT");
ax_icp_LE.set_xlabel('Fiducial Marker (index)',fontsize= 20);
ax_icp_LE.set_ylabel('Error (meters)',fontsize= 20);
ax_icp_LE.legend(loc='best', frameon=False);
# ax_icp_LE.set_title('ICP LE')
fname = trials_root + "/GEO_results/ICP_LE_" + desc_name + ".pdf"
fig_icp_LE.savefig(fname, transparent=True, pad_inches=5)
# axT.set_xlim((0,505) );
# axT.set_yticks(np.arange(0.0,250.0,20));
# # axT.legend(bbox_to_anchor=(0., 1.02, 1., .102), loc=3,
# # ncol=4, mode="expand", borderaxespad=0.)
#
# figT.savefig("/Users/isa/Experiments/reg3d_eval/downtown_dan/T_error.pdf", transparent=True, pad_inches=5)
# plt.show();
# import pdb; pdb.set_trace()
def setlogging(logfile=None):
level = logging.DEBUG
logger = logging.getLogger(__name__)
logger.setLevel(level)
# create formatter and add it to the handlers
formatter = logging.Formatter("%(asctime)s - %(name)s - %(levelname)s - %(message)s")
# create console handler with a higher log level
ch = logging.StreamHandler()
ch.setLevel(level)
ch.setFormatter(formatter)
# add the handlers to logger
logger.addHandler(ch)
# create file handler which logs error messages
if logfile:
print "Logging to file"
fh = logging.FileHandler(logfile)
fh.setLevel(level)
fh.setFormatter(formatter)
logger.addHandler(fh)
#test logging
logger.debug("debug message")
logger.info("info message")
logger.warn("warn message")
logger.error("error message")
logger.critical("critical message")
return logger
if __name__ == '__main__':
# initialize the parser object:
parser = argparse.ArgumentParser(description="Export PLY to PCD file")
# define options here:
parser.add_argument("-v", "--verbose", action='store', type = bool, dest="verbose", default=True, help="Write debug log to log_file")
parser.add_argument("-L", "--log", dest="logfile", help="write debug log to log_file")
args = parser.parse_args(argv)
# set up logging
if args.verbose:
status = main(args.logfile)
else:
status = main()
sys.exit(status)
| bsd-2-clause |
MechCoder/scikit-learn | examples/bicluster/plot_spectral_biclustering.py | 403 | 2011 | """
=============================================
A demo of the Spectral Biclustering algorithm
=============================================
This example demonstrates how to generate a checkerboard dataset and
bicluster it using the Spectral Biclustering algorithm.
The data is generated with the ``make_checkerboard`` function, then
shuffled and passed to the Spectral Biclustering algorithm. The rows
and columns of the shuffled matrix are rearranged to show the
biclusters found by the algorithm.
The outer product of the row and column label vectors shows a
representation of the checkerboard structure.
"""
print(__doc__)
# Author: Kemal Eren <kemal@kemaleren.com>
# License: BSD 3 clause
import numpy as np
from matplotlib import pyplot as plt
from sklearn.datasets import make_checkerboard
from sklearn.datasets import samples_generator as sg
from sklearn.cluster.bicluster import SpectralBiclustering
from sklearn.metrics import consensus_score
n_clusters = (4, 3)
data, rows, columns = make_checkerboard(
shape=(300, 300), n_clusters=n_clusters, noise=10,
shuffle=False, random_state=0)
plt.matshow(data, cmap=plt.cm.Blues)
plt.title("Original dataset")
data, row_idx, col_idx = sg._shuffle(data, random_state=0)
plt.matshow(data, cmap=plt.cm.Blues)
plt.title("Shuffled dataset")
model = SpectralBiclustering(n_clusters=n_clusters, method='log',
random_state=0)
model.fit(data)
score = consensus_score(model.biclusters_,
(rows[:, row_idx], columns[:, col_idx]))
print("consensus score: {:.1f}".format(score))
fit_data = data[np.argsort(model.row_labels_)]
fit_data = fit_data[:, np.argsort(model.column_labels_)]
plt.matshow(fit_data, cmap=plt.cm.Blues)
plt.title("After biclustering; rearranged to show biclusters")
plt.matshow(np.outer(np.sort(model.row_labels_) + 1,
np.sort(model.column_labels_) + 1),
cmap=plt.cm.Blues)
plt.title("Checkerboard structure of rearranged data")
plt.show()
| bsd-3-clause |
KDD-OpenSource/geox-young-academy | day-3/Kalman-filter_Mark.py | 1 | 1494 | # -*- coding: utf-8 -*-
"""
Created on Wed Oct 11 10:10:24 2017
@author: Mark
"""
import numpy as np
import matplotlib.pyplot as plt
#Define functions
def model(state_0,A,B):
state_1 = A*state_0 + np.random.normal(0,B)
return state_1
state_null=np.random.normal(0,0.4)
def observation_function(state,R):
obs=state+np.random.normal(0,R)
return obs
def forecast(state_0,cov_0,A,B):
state_1=A*state_0
cov_1=A*cov_0*A+B
return state_1,cov_1
def analysis_formulas(state_1_hat,cov_1_hat,K,H,obs_0):
state_1 = state_1_hat - K*(H*state_1_hat - obs_0)
cov_1 = cov_1_hat - K*H*cov_1_hat
return state_1, cov_1
def kalman_gain(cov_1_hat,H,R):
K = cov_1_hat*H*(R+H*cov_1_hat*H)**(-1)
return K
#Initialize model parameters
A = 0.5
H = 1
B = 0.5
R = 0.1
lev = 100
#Sythetic Model
STATE_real = np.zeros(lev)
OBS_real = np.zeros(lev)
STATE_real[0] = np.random.normal(5,0.1)
OBS_real[0] = observation_function(STATE_real[0],R)
for i in range (1,lev-1):
STATE_real[i] = model(STATE_real[i-1],0.4,0.01)
OBS_real[i] = observation_function(STATE_real[i],R)
#Kalman-filter
STATE = np.zeros(lev)
COV = np.zeros(lev)
STATE[0] = state_null
COV[0] = B
for i in range (1,lev-1):
(state_hat,cov_hat) = forecast(STATE[i-1],COV[i-1],A,B)
K = kalman_gain(cov_hat,H,R)
(STATE[i],COV[i]) = analysis_formulas(state_hat,cov_hat,K,H,OBS_real[i])
plt.plot(STATE)
plt.plot(STATE_real)
| mit |
mskwark/PconsC3 | extra/arne/MSA/find-intradom.py | 1 | 1381 | #!/usr/bin/env perl
# Find all contacts beween domains..
import sys, os, re, string
import argparse
from os.path import expanduser
home = expanduser("~")
sys.path.append(home + '/bioinfo-toolbox/parsing')
sys.path.append(home + '/git/bioinfo-toolbox/parsing')
import parse_contacts
import numpy as np
import matplotlib
matplotlib.use('Agg')
sep=5
contacts = parse_contacts.parse(open(c_filename, 'r'), sep)
contacts_np = parse_contacts.get_numpy_cmap(contacts)
contacts_np = contacts_np[start:end,start:end]
for i in range(len(contacts)):
score = contacts[i][0]
c_x = contacts[i][1] - 1
c_y = contacts[i][2] - 1
# only look at contacts within given range
# default: take full sequence range into account
if c_x < start or c_x >= end:
continue
if c_y < start or c_y >= end:
continue
if c_y-c_x < start or c_y >= end:
continue
if c_x < domain
pos_diff = abs(c_x - c_y)
too_close = pos_diff < 5
if __name__ == "__main__":
p = argparse.ArgumentParser(description='Plot protein residue contact maps.')
p.add_argument('-t', '--threshold', default=-1, type=float)
p.add_argument('--start', default=0, type=int)
p.add_argument('--end', default=-1, type=int)
p.add_argument('--sep', default=5, type=int)
p.add_argument('--domain', default=-1, type=int)
| gpl-2.0 |
goulu/Goulib | Goulib/plot.py | 1 | 4898 | """
plotable rich object display on IPython/Jupyter notebooks
"""
__author__ = "Philippe Guglielmetti"
__copyright__ = "Copyright 2015, Philippe Guglielmetti"
__credits__ = []
__license__ = "LGPL"
# import matplotlib and set backend once for all
from . import itertools2
import os
import io
import sys
import logging
import base64
import matplotlib
if os.getenv('TRAVIS'): # are we running https://travis-ci.org/ automated tests ?
matplotlib.use('Agg') # Force matplotlib not to use any Xwindows backend
elif sys.gettrace(): # http://stackoverflow.com/questions/333995/how-to-detect-that-python-code-is-being-executed-through-the-debugger
matplotlib.use('Agg') # because 'QtAgg' crashes python while debugging
else:
pass
# matplotlib.use('pdf') #for high quality pdf, but doesn't work for png, svg ...
logging.info('matplotlib backend is %s' % matplotlib.get_backend())
class Plot(object):
"""base class for plotable rich object display on IPython notebooks
inspired from http://nbviewer.ipython.org/github/ipython/ipython/blob/3607712653c66d63e0d7f13f073bde8c0f209ba8/docs/examples/notebooks/display_protocol.ipynb
"""
def _plot(self, ax, **kwargs):
"""abstract method, must be overriden
:param ax: `matplotlib.axis`
:return ax: `matplotlib.axis` after plot
"""
raise NotImplementedError(
'objects derived from plot.PLot must define a _plot method')
return ax
def render(self, fmt='svg', **kwargs):
return render([self], fmt, **kwargs) # call global function
def save(self, filename, **kwargs):
return save([self], filename, **kwargs) # call global function
# for IPython notebooks
def _repr_html_(self):
"""default rich format is svg plot"""
try:
return self._repr_svg_()
except NotImplementedError:
pass
# this returns the same as _repr_png_, but is Table compatible
buffer = self.render('png')
s = base64.b64encode(buffer).decode('utf-8')
return '<img src="data:image/png;base64,%s">' % s
def html(self, **kwargs):
from IPython.display import HTML
return HTML(self._repr_html_(**kwargs))
def svg(self, **kwargs):
from IPython.display import SVG
return SVG(self._repr_svg_(**kwargs))
def _repr_svg_(self, **kwargs):
return self.render(fmt='svg', **kwargs).decode('utf-8')
def png(self, **kwargs):
from IPython.display import Image
return Image(self._repr_png_(**kwargs), embed=True)
def _repr_png_(self, **kwargs):
return self.render(fmt='png', **kwargs)
def plot(self, **kwargs):
""" renders on IPython Notebook
(alias to make usage more straightforward)
"""
return self.svg(**kwargs)
def render(plotables, fmt='svg', **kwargs):
"""renders several Plot objects"""
import matplotlib.pyplot as plt
# extract optional arguments used for rasterization
printargs, kwargs = itertools2.dictsplit(
kwargs,
['dpi', 'transparent', 'facecolor', 'background', 'figsize']
)
ylim = kwargs.pop('ylim', None)
xlim = kwargs.pop('xlim', None)
title = kwargs.pop('title', None)
fig, ax = plt.subplots()
labels = kwargs.pop('labels', [None] * len(plotables))
# slightly shift the points to make superimposed curves more visible
offset = kwargs.pop('offset', 0)
for i, obj in enumerate(plotables):
if labels[i] is None:
labels[i] = str(obj)
if not title:
try:
title = obj._repr_latex_()
# check that title can be used in matplotlib
from matplotlib.mathtext import MathTextParser
parser = MathTextParser('path').parse(title)
except Exception as e:
title = labels[i]
ax = obj._plot(ax, label=labels[i], offset=i * offset, **kwargs)
if ylim:
plt.ylim(ylim)
if xlim:
plt.xlim(xlim)
ax.set_title(title)
if len(labels) > 1:
ax.legend()
output = io.BytesIO()
fig.savefig(output, format=fmt, **printargs)
data = output.getvalue()
plt.close(fig)
return data
def png(plotables, **kwargs):
from IPython.display import Image
return Image(render(plotables, 'png', **kwargs), embed=True)
def svg(plotables, **kwargs):
from IPython.display import SVG
return SVG(render(plotables, 'svg', **kwargs))
plot = svg
def save(plotables, filename, **kwargs):
ext = filename.split('.')[-1].lower()
kwargs.setdefault('dpi', 600) # force good quality
return open(filename, 'wb').write(render(plotables, ext, **kwargs))
| lgpl-3.0 |
elingg/tensorflow | tensorflow/contrib/learn/python/learn/learn_io/data_feeder.py | 88 | 31139 | # Copyright 2016 The TensorFlow Authors. All Rights Reserved.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
# ==============================================================================
"""Implementations of different data feeders to provide data for TF trainer."""
# TODO(ipolosukhin): Replace this module with feed-dict queue runners & queues.
from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
import itertools
import math
import numpy as np
import six
from six.moves import xrange # pylint: disable=redefined-builtin
from tensorflow.python.framework import dtypes
from tensorflow.python.framework import ops
from tensorflow.python.ops import array_ops
from tensorflow.python.platform import tf_logging as logging
# pylint: disable=g-multiple-import,g-bad-import-order
from .pandas_io import HAS_PANDAS, extract_pandas_data, extract_pandas_matrix, extract_pandas_labels
from .dask_io import HAS_DASK, extract_dask_data, extract_dask_labels
# pylint: enable=g-multiple-import,g-bad-import-order
def _get_in_out_shape(x_shape, y_shape, n_classes, batch_size=None):
"""Returns shape for input and output of the data feeder."""
x_is_dict, y_is_dict = isinstance(
x_shape, dict), y_shape is not None and isinstance(y_shape, dict)
if y_is_dict and n_classes is not None:
assert (isinstance(n_classes, dict))
if batch_size is None:
batch_size = list(x_shape.values())[0][0] if x_is_dict else x_shape[0]
elif batch_size <= 0:
raise ValueError('Invalid batch_size %d.' % batch_size)
if x_is_dict:
input_shape = {}
for k, v in list(x_shape.items()):
input_shape[k] = [batch_size] + (list(v[1:]) if len(v) > 1 else [1])
else:
x_shape = list(x_shape[1:]) if len(x_shape) > 1 else [1]
input_shape = [batch_size] + x_shape
if y_shape is None:
return input_shape, None, batch_size
def out_el_shape(out_shape, num_classes):
out_shape = list(out_shape[1:]) if len(out_shape) > 1 else []
# Skip first dimension if it is 1.
if out_shape and out_shape[0] == 1:
out_shape = out_shape[1:]
if num_classes is not None and num_classes > 1:
return [batch_size] + out_shape + [num_classes]
else:
return [batch_size] + out_shape
if not y_is_dict:
output_shape = out_el_shape(y_shape, n_classes)
else:
output_shape = dict([
(k, out_el_shape(v, n_classes[k]
if n_classes is not None and k in n_classes else None))
for k, v in list(y_shape.items())
])
return input_shape, output_shape, batch_size
def _data_type_filter(x, y):
"""Filter data types into acceptable format."""
if HAS_DASK:
x = extract_dask_data(x)
if y is not None:
y = extract_dask_labels(y)
if HAS_PANDAS:
x = extract_pandas_data(x)
if y is not None:
y = extract_pandas_labels(y)
return x, y
def _is_iterable(x):
return hasattr(x, 'next') or hasattr(x, '__next__')
def setup_train_data_feeder(x,
y,
n_classes,
batch_size=None,
shuffle=True,
epochs=None):
"""Create data feeder, to sample inputs from dataset.
If `x` and `y` are iterators, use `StreamingDataFeeder`.
Args:
x: numpy, pandas or Dask matrix or dictionary of aforementioned. Also
supports iterables.
y: numpy, pandas or Dask array or dictionary of aforementioned. Also
supports
iterables.
n_classes: number of classes. Must be None or same type as y. In case, `y`
is `dict`
(or iterable which returns dict) such that `n_classes[key] = n_classes for
y[key]`
batch_size: size to split data into parts. Must be >= 1.
shuffle: Whether to shuffle the inputs.
epochs: Number of epochs to run.
Returns:
DataFeeder object that returns training data.
Raises:
ValueError: if one of `x` and `y` is iterable and the other is not.
"""
x, y = _data_type_filter(x, y)
if HAS_DASK:
# pylint: disable=g-import-not-at-top
import dask.dataframe as dd
if (isinstance(x, (dd.Series, dd.DataFrame)) and
(y is None or isinstance(y, (dd.Series, dd.DataFrame)))):
data_feeder_cls = DaskDataFeeder
else:
data_feeder_cls = DataFeeder
else:
data_feeder_cls = DataFeeder
if _is_iterable(x):
if y is not None and not _is_iterable(y):
raise ValueError('Both x and y should be iterators for '
'streaming learning to work.')
return StreamingDataFeeder(x, y, n_classes, batch_size)
return data_feeder_cls(
x, y, n_classes, batch_size, shuffle=shuffle, epochs=epochs)
def _batch_data(x, batch_size=None):
if (batch_size is not None) and (batch_size <= 0):
raise ValueError('Invalid batch_size %d.' % batch_size)
x_first_el = six.next(x)
x = itertools.chain([x_first_el], x)
chunk = dict([(k, []) for k in list(x_first_el.keys())]) if isinstance(
x_first_el, dict) else []
chunk_filled = False
for data in x:
if isinstance(data, dict):
for k, v in list(data.items()):
chunk[k].append(v)
if (batch_size is not None) and (len(chunk[k]) >= batch_size):
chunk[k] = np.matrix(chunk[k])
chunk_filled = True
if chunk_filled:
yield chunk
chunk = dict([(k, []) for k in list(x_first_el.keys())]) if isinstance(
x_first_el, dict) else []
chunk_filled = False
else:
chunk.append(data)
if (batch_size is not None) and (len(chunk) >= batch_size):
yield np.matrix(chunk)
chunk = []
if isinstance(x_first_el, dict):
for k, v in list(data.items()):
chunk[k] = np.matrix(chunk[k])
yield chunk
else:
yield np.matrix(chunk)
def setup_predict_data_feeder(x, batch_size=None):
"""Returns an iterable for feeding into predict step.
Args:
x: numpy, pandas, Dask array or dictionary of aforementioned. Also supports
iterable.
batch_size: Size of batches to split data into. If `None`, returns one
batch of full size.
Returns:
List or iterator (or dictionary thereof) of parts of data to predict on.
Raises:
ValueError: if `batch_size` <= 0.
"""
if HAS_DASK:
x = extract_dask_data(x)
if HAS_PANDAS:
x = extract_pandas_data(x)
if _is_iterable(x):
return _batch_data(x, batch_size)
if len(x.shape) == 1:
x = np.reshape(x, (-1, 1))
if batch_size is not None:
if batch_size <= 0:
raise ValueError('Invalid batch_size %d.' % batch_size)
n_batches = int(math.ceil(float(len(x)) / batch_size))
return [x[i * batch_size:(i + 1) * batch_size] for i in xrange(n_batches)]
return [x]
def setup_processor_data_feeder(x):
"""Sets up processor iterable.
Args:
x: numpy, pandas or iterable.
Returns:
Iterable of data to process.
"""
if HAS_PANDAS:
x = extract_pandas_matrix(x)
return x
def check_array(array, dtype):
"""Checks array on dtype and converts it if different.
Args:
array: Input array.
dtype: Expected dtype.
Returns:
Original array or converted.
"""
# skip check if array is instance of other classes, e.g. h5py.Dataset
# to avoid copying array and loading whole data into memory
if isinstance(array, (np.ndarray, list)):
array = np.array(array, dtype=dtype, order=None, copy=False)
return array
def _access(data, iloc):
"""Accesses an element from collection, using integer location based indexing.
Args:
data: array-like. The collection to access
iloc: `int` or `list` of `int`s. Location(s) to access in `collection`
Returns:
The element of `a` found at location(s) `iloc`.
"""
if HAS_PANDAS:
import pandas as pd # pylint: disable=g-import-not-at-top
if isinstance(data, pd.Series) or isinstance(data, pd.DataFrame):
return data.iloc[iloc]
return data[iloc]
def _check_dtype(dtype):
if dtypes.as_dtype(dtype) == dtypes.float64:
logging.warn(
'float64 is not supported by many models, consider casting to float32.')
return dtype
class DataFeeder(object):
"""Data feeder is an example class to sample data for TF trainer."""
def __init__(self,
x,
y,
n_classes,
batch_size=None,
shuffle=True,
random_state=None,
epochs=None):
"""Initializes a DataFeeder instance.
Args:
x: One feature sample which can either Nd numpy matrix of shape
`[n_samples, n_features, ...]` or dictionary of Nd numpy matrix.
y: label vector, either floats for regression or class id for
classification. If matrix, will consider as a sequence of labels.
Can be `None` for unsupervised setting. Also supports dictionary of
labels.
n_classes: Number of classes, 0 and 1 are considered regression, `None`
will pass through the input labels without one-hot conversion. Also, if
`y` is `dict`, then `n_classes` must be `dict` such that
`n_classes[key] = n_classes for label y[key]`, `None` otherwise.
batch_size: Mini-batch size to accumulate samples in one mini batch.
shuffle: Whether to shuffle `x`.
random_state: Numpy `RandomState` object to reproduce sampling.
epochs: Number of times to iterate over input data before raising
`StopIteration` exception.
Attributes:
x: Input features (ndarray or dictionary of ndarrays).
y: Input label (ndarray or dictionary of ndarrays).
n_classes: Number of classes (if `None`, pass through indices without
one-hot conversion).
batch_size: Mini-batch size to accumulate.
input_shape: Shape of the input (or dictionary of shapes).
output_shape: Shape of the output (or dictionary of shapes).
input_dtype: DType of input (or dictionary of shapes).
output_dtype: DType of output (or dictionary of shapes.
"""
x_is_dict, y_is_dict = isinstance(x, dict), y is not None and isinstance(
y, dict)
if isinstance(y, list):
y = np.array(y)
self._x = dict([(k, check_array(v, v.dtype)) for k, v in list(x.items())
]) if x_is_dict else check_array(x, x.dtype)
self._y = None if y is None else \
dict([(k, check_array(v, v.dtype)) for k, v in list(y.items())]) if x_is_dict else check_array(y, y.dtype)
# self.n_classes is not None means we're converting raw target indices to one-hot.
if n_classes is not None:
if not y_is_dict:
y_dtype = (np.int64
if n_classes is not None and n_classes > 1 else np.float32)
self._y = (None if y is None else check_array(y, dtype=y_dtype))
self.n_classes = n_classes
self.max_epochs = epochs
x_shape = dict([(k, v.shape) for k, v in list(self._x.items())
]) if x_is_dict else self._x.shape
y_shape = dict([(k, v.shape) for k, v in list(self._y.items())
]) if y_is_dict else None if y is None else self._y.shape
self.input_shape, self.output_shape, self._batch_size = _get_in_out_shape(
x_shape, y_shape, n_classes, batch_size)
# Input dtype matches dtype of x.
self._input_dtype = dict([(k, _check_dtype(v.dtype)) for k, v in list(self._x.items())]) if x_is_dict \
else _check_dtype(self._x.dtype)
# note: self._output_dtype = np.float32 when y is None
self._output_dtype = dict([(k, _check_dtype(v.dtype)) for k, v in list(self._y.items())]) if y_is_dict \
else _check_dtype(self._y.dtype) if y is not None else np.float32
# self.n_classes is None means we're passing in raw target indices
if n_classes is not None and y_is_dict:
for key in list(n_classes.keys()):
if key in self._output_dtype:
self._output_dtype[key] = np.float32
self._shuffle = shuffle
self.random_state = np.random.RandomState(
42) if random_state is None else random_state
num_samples = list(self._x.values())[0].shape[
0] if x_is_dict else self._x.shape[0]
if self._shuffle:
self.indices = self.random_state.permutation(num_samples)
else:
self.indices = np.array(range(num_samples))
self.offset = 0
self.epoch = 0
self._epoch_placeholder = None
@property
def x(self):
return self._x
@property
def y(self):
return self._y
@property
def shuffle(self):
return self._shuffle
@property
def input_dtype(self):
return self._input_dtype
@property
def output_dtype(self):
return self._output_dtype
@property
def batch_size(self):
return self._batch_size
def make_epoch_variable(self):
"""Adds a placeholder variable for the epoch to the graph.
Returns:
The epoch placeholder.
"""
self._epoch_placeholder = array_ops.placeholder(
dtypes.int32, [1], name='epoch')
return self._epoch_placeholder
def input_builder(self):
"""Builds inputs in the graph.
Returns:
Two placeholders for inputs and outputs.
"""
def get_placeholder(shape, dtype, name_prepend):
if shape is None:
return None
if isinstance(shape, dict):
placeholder = {}
for key in list(shape.keys()):
placeholder[key] = array_ops.placeholder(
dtypes.as_dtype(dtype[key]), [None] + shape[key][1:],
name=name_prepend + '_' + key)
else:
placeholder = array_ops.placeholder(
dtypes.as_dtype(dtype), [None] + shape[1:], name=name_prepend)
return placeholder
self._input_placeholder = get_placeholder(self.input_shape,
self._input_dtype, 'input')
self._output_placeholder = get_placeholder(self.output_shape,
self._output_dtype, 'output')
return self._input_placeholder, self._output_placeholder
def set_placeholders(self, input_placeholder, output_placeholder):
"""Sets placeholders for this data feeder.
Args:
input_placeholder: Placeholder for `x` variable. Should match shape
of the examples in the x dataset.
output_placeholder: Placeholder for `y` variable. Should match
shape of the examples in the y dataset. Can be `None`.
"""
self._input_placeholder = input_placeholder
self._output_placeholder = output_placeholder
def get_feed_params(self):
"""Function returns a `dict` with data feed params while training.
Returns:
A `dict` with data feed params while training.
"""
return {
'epoch': self.epoch,
'offset': self.offset,
'batch_size': self._batch_size
}
def get_feed_dict_fn(self):
"""Returns a function that samples data into given placeholders.
Returns:
A function that when called samples a random subset of batch size
from `x` and `y`.
"""
x_is_dict, y_is_dict = isinstance(
self._x, dict), self._y is not None and isinstance(self._y, dict)
# Assign input features from random indices.
def extract(data, indices):
return (np.array(_access(data, indices)).reshape((indices.shape[0], 1)) if
len(data.shape) == 1 else _access(data, indices))
# assign labels from random indices
def assign_label(data, shape, dtype, n_classes, indices):
shape[0] = indices.shape[0]
out = np.zeros(shape, dtype=dtype)
for i in xrange(out.shape[0]):
sample = indices[i]
# self.n_classes is None means we're passing in raw target indices
if n_classes is None:
out[i] = _access(data, sample)
else:
if n_classes > 1:
if len(shape) == 2:
out.itemset((i, int(_access(data, sample))), 1.0)
else:
for idx, value in enumerate(_access(data, sample)):
out.itemset(tuple([i, idx, value]), 1.0)
else:
out[i] = _access(data, sample)
return out
def _feed_dict_fn():
"""Function that samples data into given placeholders."""
if self.max_epochs is not None and self.epoch + 1 > self.max_epochs:
raise StopIteration
assert self._input_placeholder is not None
feed_dict = {}
if self._epoch_placeholder is not None:
feed_dict[self._epoch_placeholder.name] = [self.epoch]
# Take next batch of indices.
x_len = list(self._x.values())[0].shape[
0] if x_is_dict else self._x.shape[0]
end = min(x_len, self.offset + self._batch_size)
batch_indices = self.indices[self.offset:end]
# adding input placeholder
feed_dict.update(
dict([(self._input_placeholder[k].name, extract(v, batch_indices))
for k, v in list(self._x.items())]) if x_is_dict else
{self._input_placeholder.name: extract(self._x, batch_indices)})
# move offset and reset it if necessary
self.offset += self._batch_size
if self.offset >= x_len:
self.indices = self.random_state.permutation(
x_len) if self._shuffle else np.array(range(x_len))
self.offset = 0
self.epoch += 1
# return early if there are no labels
if self._output_placeholder is None:
return feed_dict
# adding output placeholders
if y_is_dict:
for k, v in list(self._y.items()):
n_classes = (self.n_classes[k] if k in self.n_classes else
None) if self.n_classes is not None else None
shape, dtype = self.output_shape[k], self._output_dtype[k]
feed_dict.update({
self._output_placeholder[k].name:
assign_label(v, shape, dtype, n_classes, batch_indices)
})
else:
shape, dtype, n_classes = self.output_shape, self._output_dtype, self.n_classes
feed_dict.update({
self._output_placeholder.name:
assign_label(self._y, shape, dtype, n_classes, batch_indices)
})
return feed_dict
return _feed_dict_fn
class StreamingDataFeeder(DataFeeder):
"""Data feeder for TF trainer that reads data from iterator.
Streaming data feeder allows to read data as it comes it from disk or
somewhere else. It's custom to have this iterators rotate infinetly over
the dataset, to allow control of how much to learn on the trainer side.
"""
def __init__(self, x, y, n_classes, batch_size):
"""Initializes a StreamingDataFeeder instance.
Args:
x: iterator each element of which returns one feature sample. Sample can
be a Nd numpy matrix or dictionary of Nd numpy matrices.
y: iterator each element of which returns one label sample. Sample can be
a Nd numpy matrix or dictionary of Nd numpy matrices with 1 or many
classes regression values.
n_classes: indicator of how many classes the corresponding label sample
has for the purposes of one-hot conversion of label. In case where `y`
is a dictionary, `n_classes` must be dictionary (with same keys as `y`)
of how many classes there are in each label in `y`. If key is
present in `y` and missing in `n_classes`, the value is assumed `None`
and no one-hot conversion will be applied to the label with that key.
batch_size: Mini batch size to accumulate samples in one batch. If set
`None`, then assumes that iterator to return already batched element.
Attributes:
x: input features (or dictionary of input features).
y: input label (or dictionary of output features).
n_classes: number of classes.
batch_size: mini batch size to accumulate.
input_shape: shape of the input (can be dictionary depending on `x`).
output_shape: shape of the output (can be dictionary depending on `y`).
input_dtype: dtype of input (can be dictionary depending on `x`).
output_dtype: dtype of output (can be dictionary depending on `y`).
"""
# pylint: disable=invalid-name,super-init-not-called
x_first_el = six.next(x)
self._x = itertools.chain([x_first_el], x)
if y is not None:
y_first_el = six.next(y)
self._y = itertools.chain([y_first_el], y)
else:
y_first_el = None
self._y = None
self.n_classes = n_classes
x_is_dict = isinstance(x_first_el, dict)
y_is_dict = y is not None and isinstance(y_first_el, dict)
if y_is_dict and n_classes is not None:
assert isinstance(n_classes, dict)
# extract shapes for first_elements
if x_is_dict:
x_first_el_shape = dict(
[(k, [1] + list(v.shape)) for k, v in list(x_first_el.items())])
else:
x_first_el_shape = [1] + list(x_first_el.shape)
if y_is_dict:
y_first_el_shape = dict(
[(k, [1] + list(v.shape)) for k, v in list(y_first_el.items())])
elif y is None:
y_first_el_shape = None
else:
y_first_el_shape = ([1] + list(y_first_el[0].shape if isinstance(
y_first_el, list) else y_first_el.shape))
self.input_shape, self.output_shape, self._batch_size = _get_in_out_shape(
x_first_el_shape, y_first_el_shape, n_classes, batch_size)
# Input dtype of x_first_el.
if x_is_dict:
self._input_dtype = dict(
[(k, _check_dtype(v.dtype)) for k, v in list(x_first_el.items())])
else:
self._input_dtype = _check_dtype(x_first_el.dtype)
# Output dtype of y_first_el.
def check_y_dtype(el):
if isinstance(el, np.ndarray):
return el.dtype
elif isinstance(el, list):
return check_y_dtype(el[0])
else:
return _check_dtype(np.dtype(type(el)))
# Output types are floats, due to both softmaxes and regression req.
if n_classes is not None and (y is None or not y_is_dict) and n_classes > 0:
self._output_dtype = np.float32
elif y_is_dict:
self._output_dtype = dict(
[(k, check_y_dtype(v)) for k, v in list(y_first_el.items())])
elif y is None:
self._output_dtype = None
else:
self._output_dtype = check_y_dtype(y_first_el)
def get_feed_params(self):
"""Function returns a `dict` with data feed params while training.
Returns:
A `dict` with data feed params while training.
"""
return {'batch_size': self._batch_size}
def get_feed_dict_fn(self):
"""Returns a function, that will sample data and provide it to placeholders.
Returns:
A function that when called samples a random subset of batch size
from x and y.
"""
self.stopped = False
def _feed_dict_fn():
"""Samples data and provides it to placeholders.
Returns:
`dict` of input and output tensors.
"""
def init_array(shape, dtype):
"""Initialize array of given shape or dict of shapes and dtype."""
if shape is None:
return None
elif isinstance(shape, dict):
return dict([(k, np.zeros(shape[k], dtype[k]))
for k in list(shape.keys())])
else:
return np.zeros(shape, dtype=dtype)
def put_data_array(dest, index, source=None, n_classes=None):
"""Puts data array into container."""
if source is None:
dest = dest[:index]
elif n_classes is not None and n_classes > 1:
if len(self.output_shape) == 2:
dest.itemset((index, source), 1.0)
else:
for idx, value in enumerate(source):
dest.itemset(tuple([index, idx, value]), 1.0)
else:
if len(dest.shape) > 1:
dest[index, :] = source
else:
dest[index] = source[0] if isinstance(source, list) else source
return dest
def put_data_array_or_dict(holder, index, data=None, n_classes=None):
"""Puts data array or data dictionary into container."""
if holder is None:
return None
if isinstance(holder, dict):
if data is None:
data = {k: None for k in holder.keys()}
assert isinstance(data, dict)
for k in holder.keys():
num_classes = n_classes[k] if (n_classes is not None and
k in n_classes) else None
holder[k] = put_data_array(holder[k], index, data[k], num_classes)
else:
holder = put_data_array(holder, index, data, n_classes)
return holder
if self.stopped:
raise StopIteration
inp = init_array(self.input_shape, self._input_dtype)
out = init_array(self.output_shape, self._output_dtype)
for i in xrange(self._batch_size):
# Add handling when queue ends.
try:
next_inp = six.next(self._x)
inp = put_data_array_or_dict(inp, i, next_inp, None)
except StopIteration:
self.stopped = True
if i == 0:
raise
inp = put_data_array_or_dict(inp, i, None, None)
out = put_data_array_or_dict(out, i, None, None)
break
if self._y is not None:
next_out = six.next(self._y)
out = put_data_array_or_dict(out, i, next_out, self.n_classes)
# creating feed_dict
if isinstance(inp, dict):
feed_dict = dict([(self._input_placeholder[k].name, inp[k])
for k in list(self._input_placeholder.keys())])
else:
feed_dict = {self._input_placeholder.name: inp}
if self._y is not None:
if isinstance(out, dict):
feed_dict.update(
dict([(self._output_placeholder[k].name, out[k])
for k in list(self._output_placeholder.keys())]))
else:
feed_dict.update({self._output_placeholder.name: out})
return feed_dict
return _feed_dict_fn
class DaskDataFeeder(object):
"""Data feeder for that reads data from dask.Series and dask.DataFrame.
Numpy arrays can be serialized to disk and it's possible to do random seeks
into them. DaskDataFeeder will remove requirement to have full dataset in the
memory and still do random seeks for sampling of batches.
"""
def __init__(self,
x,
y,
n_classes,
batch_size,
shuffle=True,
random_state=None,
epochs=None):
"""Initializes a DaskDataFeeder instance.
Args:
x: iterator that returns for each element, returns features.
y: iterator that returns for each element, returns 1 or many classes /
regression values.
n_classes: indicator of how many classes the label has.
batch_size: Mini batch size to accumulate.
shuffle: Whether to shuffle the inputs.
random_state: random state for RNG. Note that it will mutate so use a
int value for this if you want consistent sized batches.
epochs: Number of epochs to run.
Attributes:
x: input features.
y: input label.
n_classes: number of classes.
batch_size: mini batch size to accumulate.
input_shape: shape of the input.
output_shape: shape of the output.
input_dtype: dtype of input.
output_dtype: dtype of output.
Raises:
ValueError: if `x` or `y` are `dict`, as they are not supported currently.
"""
if isinstance(x, dict) or isinstance(y, dict):
raise ValueError(
'DaskDataFeeder does not support dictionaries at the moment.')
# pylint: disable=invalid-name,super-init-not-called
import dask.dataframe as dd # pylint: disable=g-import-not-at-top
# TODO(terrytangyuan): check x and y dtypes in dask_io like pandas
self._x = x
self._y = y
# save column names
self._x_columns = list(x.columns)
if isinstance(y.columns[0], str):
self._y_columns = list(y.columns)
else:
# deal with cases where two DFs have overlapped default numeric colnames
self._y_columns = len(self._x_columns) + 1
self._y = self._y.rename(columns={y.columns[0]: self._y_columns})
# TODO(terrytangyuan): deal with unsupervised cases
# combine into a data frame
self.df = dd.multi.concat([self._x, self._y], axis=1)
self.n_classes = n_classes
x_count = x.count().compute()[0]
x_shape = (x_count, len(self._x.columns))
y_shape = (x_count, len(self._y.columns))
# TODO(terrytangyuan): Add support for shuffle and epochs.
self._shuffle = shuffle
self.epochs = epochs
self.input_shape, self.output_shape, self._batch_size = _get_in_out_shape(
x_shape, y_shape, n_classes, batch_size)
self.sample_fraction = self._batch_size / float(x_count)
self._input_dtype = _check_dtype(self._x.dtypes[0])
self._output_dtype = _check_dtype(self._y.dtypes[self._y_columns])
if random_state is None:
self.random_state = 66
else:
self.random_state = random_state
def get_feed_params(self):
"""Function returns a `dict` with data feed params while training.
Returns:
A `dict` with data feed params while training.
"""
return {'batch_size': self._batch_size}
def get_feed_dict_fn(self, input_placeholder, output_placeholder):
"""Returns a function, that will sample data and provide it to placeholders.
Args:
input_placeholder: tf.Placeholder for input features mini batch.
output_placeholder: tf.Placeholder for output labels.
Returns:
A function that when called samples a random subset of batch size
from x and y.
"""
def _feed_dict_fn():
"""Samples data and provides it to placeholders."""
# TODO(ipolosukhin): option for with/without replacement (dev version of
# dask)
sample = self.df.random_split(
[self.sample_fraction, 1 - self.sample_fraction],
random_state=self.random_state)
inp = extract_pandas_matrix(sample[0][self._x_columns].compute()).tolist()
out = extract_pandas_matrix(sample[0][self._y_columns].compute())
# convert to correct dtype
inp = np.array(inp, dtype=self._input_dtype)
# one-hot encode out for each class for cross entropy loss
if HAS_PANDAS:
import pandas as pd # pylint: disable=g-import-not-at-top
if not isinstance(out, pd.Series):
out = out.flatten()
out_max = self._y.max().compute().values[0]
encoded_out = np.zeros((out.size, out_max + 1), dtype=self._output_dtype)
encoded_out[np.arange(out.size), out] = 1
return {input_placeholder.name: inp, output_placeholder.name: encoded_out}
return _feed_dict_fn
| apache-2.0 |
yavalvas/yav_com | build/matplotlib/lib/mpl_toolkits/axes_grid1/mpl_axes.py | 8 | 4971 | from __future__ import (absolute_import, division, print_function,
unicode_literals)
import six
import warnings
import matplotlib.axes as maxes
from matplotlib.artist import Artist
from matplotlib.axis import XAxis, YAxis
class SimpleChainedObjects(object):
def __init__(self, objects):
self._objects = objects
def __getattr__(self, k):
_a = SimpleChainedObjects([getattr(a, k) for a in self._objects])
return _a
def __call__(self, *kl, **kwargs):
for m in self._objects:
m(*kl, **kwargs)
class Axes(maxes.Axes):
def toggle_axisline(self, b):
warnings.warn("toggle_axisline is not necessary and deprecated in axes_grid1")
class AxisDict(dict):
def __init__(self, axes):
self.axes = axes
super(Axes.AxisDict, self).__init__()
def __getitem__(self, k):
if isinstance(k, tuple):
r = SimpleChainedObjects([dict.__getitem__(self, k1) for k1 in k])
return r
elif isinstance(k, slice):
if k.start == None and k.stop == None and k.step == None:
r = SimpleChainedObjects(list(six.itervalues(self)))
return r
else:
raise ValueError("Unsupported slice")
else:
return dict.__getitem__(self, k)
def __call__(self, *v, **kwargs):
return maxes.Axes.axis(self.axes, *v, **kwargs)
def __init__(self, *kl, **kw):
super(Axes, self).__init__(*kl, **kw)
def _init_axis_artists(self, axes=None):
if axes is None:
axes = self
self._axislines = self.AxisDict(self)
self._axislines["bottom"] = SimpleAxisArtist(self.xaxis, 1, self.spines["bottom"])
self._axislines["top"] = SimpleAxisArtist(self.xaxis, 2, self.spines["top"])
self._axislines["left"] = SimpleAxisArtist(self.yaxis, 1, self.spines["left"])
self._axislines["right"] = SimpleAxisArtist(self.yaxis, 2, self.spines["right"])
def _get_axislines(self):
return self._axislines
axis = property(_get_axislines)
def cla(self):
super(Axes, self).cla()
self._init_axis_artists()
class SimpleAxisArtist(Artist):
def __init__(self, axis, axisnum, spine):
self._axis = axis
self._axisnum = axisnum
self.line = spine
if isinstance(axis, XAxis):
self._axis_direction = ["bottom", "top"][axisnum-1]
elif isinstance(axis, YAxis):
self._axis_direction = ["left", "right"][axisnum-1]
else:
raise ValueError("axis must be instance of XAxis or YAxis : %s is provided" % (axis,))
Artist.__init__(self)
def _get_major_ticks(self):
tickline = "tick%dline" % self._axisnum
return SimpleChainedObjects([getattr(tick, tickline) for tick \
in self._axis.get_major_ticks()])
def _get_major_ticklabels(self):
label = "label%d" % self._axisnum
return SimpleChainedObjects([getattr(tick, label) for tick \
in self._axis.get_major_ticks()])
def _get_label(self):
return self._axis.label
major_ticks = property(_get_major_ticks)
major_ticklabels = property(_get_major_ticklabels)
label = property(_get_label)
def set_visible(self, b):
self.toggle(all=b)
self.line.set_visible(b)
self._axis.set_visible(True)
Artist.set_visible(self, b)
def set_label(self, txt):
self._axis.set_label_text(txt)
def toggle(self, all=None, ticks=None, ticklabels=None, label=None):
if all:
_ticks, _ticklabels, _label = True, True, True
elif all is not None:
_ticks, _ticklabels, _label = False, False, False
else:
_ticks, _ticklabels, _label = None, None, None
if ticks is not None:
_ticks = ticks
if ticklabels is not None:
_ticklabels = ticklabels
if label is not None:
_label = label
tickOn = "tick%dOn" % self._axisnum
labelOn = "label%dOn" % self._axisnum
if _ticks is not None:
tickparam = {tickOn: _ticks}
self._axis.set_tick_params(**tickparam)
if _ticklabels is not None:
tickparam = {labelOn: _ticklabels}
self._axis.set_tick_params(**tickparam)
if _label is not None:
pos = self._axis.get_label_position()
if (pos == self._axis_direction) and not _label:
self._axis.label.set_visible(False)
elif _label:
self._axis.label.set_visible(True)
self._axis.set_label_position(self._axis_direction)
if __name__ == '__main__':
fig = figure()
ax = Axes(fig, [0.1, 0.1, 0.8, 0.8])
fig.add_axes(ax)
ax.cla()
| mit |
wavelets/zipline | tests/test_batchtransform.py | 3 | 9739 | #
# Copyright 2013 Quantopian, Inc.
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
from collections import deque
import pytz
import numpy as np
import pandas as pd
from datetime import datetime
from unittest import TestCase
from zipline.utils.test_utils import setup_logger
from zipline.sources.data_source import DataSource
import zipline.utils.factory as factory
from zipline.transforms import batch_transform
from zipline.test_algorithms import (BatchTransformAlgorithm,
BatchTransformAlgorithmMinute,
ReturnPriceBatchTransform)
from zipline.algorithm import TradingAlgorithm
from zipline.utils.tradingcalendar import trading_days
from copy import deepcopy
@batch_transform
def return_price(data):
return data.price
class BatchTransformAlgorithmSetSid(TradingAlgorithm):
def initialize(self, sids=None):
self.history = []
self.batch_transform = return_price(
refresh_period=1,
window_length=10,
clean_nans=False,
sids=sids,
compute_only_full=False
)
def handle_data(self, data):
self.history.append(
deepcopy(self.batch_transform.handle_data(data)))
class DifferentSidSource(DataSource):
def __init__(self):
self.dates = pd.date_range('1990-01-01', periods=180, tz='utc')
self.start = self.dates[0]
self.end = self.dates[-1]
self._raw_data = None
self.sids = range(90)
self.sid = 0
self.trading_days = []
@property
def instance_hash(self):
return '1234'
@property
def raw_data(self):
if not self._raw_data:
self._raw_data = self.raw_data_gen()
return self._raw_data
@property
def mapping(self):
return {
'dt': (lambda x: x, 'dt'),
'sid': (lambda x: x, 'sid'),
'price': (float, 'price'),
'volume': (int, 'volume'),
}
def raw_data_gen(self):
# Create differente sid for each event
for date in self.dates:
if date not in trading_days:
continue
event = {'dt': date,
'sid': self.sid,
'price': self.sid,
'volume': self.sid}
self.sid += 1
self.trading_days.append(date)
yield event
class TestChangeOfSids(TestCase):
def setUp(self):
self.sids = range(90)
self.sim_params = factory.create_simulation_parameters(
start=datetime(1990, 1, 1, tzinfo=pytz.utc),
end=datetime(1990, 1, 8, tzinfo=pytz.utc)
)
def test_all_sids_passed(self):
algo = BatchTransformAlgorithmSetSid(sim_params=self.sim_params)
source = DifferentSidSource()
algo.run(source)
for i, (df, date) in enumerate(zip(algo.history, source.trading_days)):
self.assertEqual(df.index[-1], date, "Newest event doesn't \
match.")
for sid in self.sids[:i]:
self.assertIn(sid, df.columns)
last_elem = len(df) - 1
self.assertEqual(df[last_elem][last_elem], last_elem)
class TestBatchTransformMinutely(TestCase):
def setUp(self):
start = pd.datetime(1990, 1, 3, 0, 0, 0, 0, pytz.utc)
end = pd.datetime(1990, 1, 8, 0, 0, 0, 0, pytz.utc)
self.sim_params = factory.create_simulation_parameters(
start=start,
end=end,
)
self.sim_params.emission_rate = 'daily'
self.sim_params.data_frequency = 'minute'
setup_logger(self)
self.source, self.df = \
factory.create_test_df_source(bars='minute')
def test_core(self):
algo = BatchTransformAlgorithmMinute(sim_params=self.sim_params)
algo.run(self.source)
wl = int(algo.window_length * 6.5 * 60)
for bt in algo.history[wl:]:
self.assertEqual(len(bt), wl)
def test_window_length(self):
algo = BatchTransformAlgorithmMinute(sim_params=self.sim_params,
window_length=1, refresh_period=0)
algo.run(self.source)
wl = int(algo.window_length * 6.5 * 60)
np.testing.assert_array_equal(algo.history[:(wl - 1)],
[None] * (wl - 1))
for bt in algo.history[wl:]:
self.assertEqual(len(bt), wl)
class TestBatchTransform(TestCase):
def setUp(self):
self.sim_params = factory.create_simulation_parameters(
start=datetime(1990, 1, 1, tzinfo=pytz.utc),
end=datetime(1990, 1, 8, tzinfo=pytz.utc)
)
setup_logger(self)
self.source, self.df = \
factory.create_test_df_source(self.sim_params)
def test_core_functionality(self):
algo = BatchTransformAlgorithm(sim_params=self.sim_params)
algo.run(self.source)
wl = algo.window_length
# The following assertion depend on window length of 3
self.assertEqual(wl, 3)
# If window_length is 3, there should be 2 None events, as the
# window fills up on the 3rd day.
n_none_events = 2
self.assertEqual(algo.history_return_price_class[:n_none_events],
[None] * n_none_events,
"First two iterations should return None." + "\n" +
"i.e. no returned values until window is full'" +
"%s" % (algo.history_return_price_class,))
self.assertEqual(algo.history_return_price_decorator[:n_none_events],
[None] * n_none_events,
"First two iterations should return None." + "\n" +
"i.e. no returned values until window is full'" +
"%s" % (algo.history_return_price_decorator,))
# After three Nones, the next value should be a data frame
self.assertTrue(isinstance(
algo.history_return_price_class[wl],
pd.DataFrame)
)
# Test whether arbitrary fields can be added to datapanel
field = algo.history_return_arbitrary_fields[-1]
self.assertTrue(
'arbitrary' in field.items,
'datapanel should contain column arbitrary'
)
self.assertTrue(all(
field['arbitrary'].values.flatten() ==
[123] * algo.window_length),
'arbitrary dataframe should contain only "test"'
)
for data in algo.history_return_sid_filter[wl:]:
self.assertIn(0, data.columns)
self.assertNotIn(1, data.columns)
for data in algo.history_return_field_filter[wl:]:
self.assertIn('price', data.items)
self.assertNotIn('ignore', data.items)
for data in algo.history_return_field_no_filter[wl:]:
self.assertIn('price', data.items)
self.assertIn('ignore', data.items)
for data in algo.history_return_ticks[wl:]:
self.assertTrue(isinstance(data, deque))
for data in algo.history_return_not_full:
self.assertIsNot(data, None)
# test overloaded class
for test_history in [algo.history_return_price_class,
algo.history_return_price_decorator]:
# starting at window length, the window should contain
# consecutive (of window length) numbers up till the end.
for i in range(algo.window_length, len(test_history)):
np.testing.assert_array_equal(
range(i - algo.window_length + 2, i + 2),
test_history[i].values.flatten()
)
def test_passing_of_args(self):
algo = BatchTransformAlgorithm(1, kwarg='str',
sim_params=self.sim_params)
self.assertEqual(algo.args, (1,))
self.assertEqual(algo.kwargs, {'kwarg': 'str'})
algo.run(self.source)
expected_item = ((1, ), {'kwarg': 'str'})
self.assertEqual(
algo.history_return_args,
[
# 1990-01-01 - market holiday, no event
# 1990-01-02 - window not full
None,
# 1990-01-03 - window not full
None,
# 1990-01-04 - window now full, 3rd event
expected_item,
# 1990-01-05 - window now full
expected_item,
# 1990-01-08 - window now full
expected_item
])
def run_batchtransform(window_length=10):
sim_params = factory.create_simulation_parameters(
start=datetime(1990, 1, 1, tzinfo=pytz.utc),
end=datetime(1995, 1, 8, tzinfo=pytz.utc)
)
source, df = factory.create_test_df_source(sim_params)
return_price_class = ReturnPriceBatchTransform(
refresh_period=1,
window_length=window_length,
clean_nans=False
)
for raw_event in source:
raw_event['datetime'] = raw_event.dt
event = {0: raw_event}
return_price_class.handle_data(event)
| apache-2.0 |
Srisai85/scikit-learn | sklearn/decomposition/tests/test_truncated_svd.py | 240 | 6055 | """Test truncated SVD transformer."""
import numpy as np
import scipy.sparse as sp
from sklearn.decomposition import TruncatedSVD
from sklearn.utils import check_random_state
from sklearn.utils.testing import (assert_array_almost_equal, assert_equal,
assert_raises, assert_greater,
assert_array_less)
# Make an X that looks somewhat like a small tf-idf matrix.
# XXX newer versions of SciPy have scipy.sparse.rand for this.
shape = 60, 55
n_samples, n_features = shape
rng = check_random_state(42)
X = rng.randint(-100, 20, np.product(shape)).reshape(shape)
X = sp.csr_matrix(np.maximum(X, 0), dtype=np.float64)
X.data[:] = 1 + np.log(X.data)
Xdense = X.A
def test_algorithms():
svd_a = TruncatedSVD(30, algorithm="arpack")
svd_r = TruncatedSVD(30, algorithm="randomized", random_state=42)
Xa = svd_a.fit_transform(X)[:, :6]
Xr = svd_r.fit_transform(X)[:, :6]
assert_array_almost_equal(Xa, Xr)
comp_a = np.abs(svd_a.components_)
comp_r = np.abs(svd_r.components_)
# All elements are equal, but some elements are more equal than others.
assert_array_almost_equal(comp_a[:9], comp_r[:9])
assert_array_almost_equal(comp_a[9:], comp_r[9:], decimal=3)
def test_attributes():
for n_components in (10, 25, 41):
tsvd = TruncatedSVD(n_components).fit(X)
assert_equal(tsvd.n_components, n_components)
assert_equal(tsvd.components_.shape, (n_components, n_features))
def test_too_many_components():
for algorithm in ["arpack", "randomized"]:
for n_components in (n_features, n_features+1):
tsvd = TruncatedSVD(n_components=n_components, algorithm=algorithm)
assert_raises(ValueError, tsvd.fit, X)
def test_sparse_formats():
for fmt in ("array", "csr", "csc", "coo", "lil"):
Xfmt = Xdense if fmt == "dense" else getattr(X, "to" + fmt)()
tsvd = TruncatedSVD(n_components=11)
Xtrans = tsvd.fit_transform(Xfmt)
assert_equal(Xtrans.shape, (n_samples, 11))
Xtrans = tsvd.transform(Xfmt)
assert_equal(Xtrans.shape, (n_samples, 11))
def test_inverse_transform():
for algo in ("arpack", "randomized"):
# We need a lot of components for the reconstruction to be "almost
# equal" in all positions. XXX Test means or sums instead?
tsvd = TruncatedSVD(n_components=52, random_state=42)
Xt = tsvd.fit_transform(X)
Xinv = tsvd.inverse_transform(Xt)
assert_array_almost_equal(Xinv, Xdense, decimal=1)
def test_integers():
Xint = X.astype(np.int64)
tsvd = TruncatedSVD(n_components=6)
Xtrans = tsvd.fit_transform(Xint)
assert_equal(Xtrans.shape, (n_samples, tsvd.n_components))
def test_explained_variance():
# Test sparse data
svd_a_10_sp = TruncatedSVD(10, algorithm="arpack")
svd_r_10_sp = TruncatedSVD(10, algorithm="randomized", random_state=42)
svd_a_20_sp = TruncatedSVD(20, algorithm="arpack")
svd_r_20_sp = TruncatedSVD(20, algorithm="randomized", random_state=42)
X_trans_a_10_sp = svd_a_10_sp.fit_transform(X)
X_trans_r_10_sp = svd_r_10_sp.fit_transform(X)
X_trans_a_20_sp = svd_a_20_sp.fit_transform(X)
X_trans_r_20_sp = svd_r_20_sp.fit_transform(X)
# Test dense data
svd_a_10_de = TruncatedSVD(10, algorithm="arpack")
svd_r_10_de = TruncatedSVD(10, algorithm="randomized", random_state=42)
svd_a_20_de = TruncatedSVD(20, algorithm="arpack")
svd_r_20_de = TruncatedSVD(20, algorithm="randomized", random_state=42)
X_trans_a_10_de = svd_a_10_de.fit_transform(X.toarray())
X_trans_r_10_de = svd_r_10_de.fit_transform(X.toarray())
X_trans_a_20_de = svd_a_20_de.fit_transform(X.toarray())
X_trans_r_20_de = svd_r_20_de.fit_transform(X.toarray())
# helper arrays for tests below
svds = (svd_a_10_sp, svd_r_10_sp, svd_a_20_sp, svd_r_20_sp, svd_a_10_de,
svd_r_10_de, svd_a_20_de, svd_r_20_de)
svds_trans = (
(svd_a_10_sp, X_trans_a_10_sp),
(svd_r_10_sp, X_trans_r_10_sp),
(svd_a_20_sp, X_trans_a_20_sp),
(svd_r_20_sp, X_trans_r_20_sp),
(svd_a_10_de, X_trans_a_10_de),
(svd_r_10_de, X_trans_r_10_de),
(svd_a_20_de, X_trans_a_20_de),
(svd_r_20_de, X_trans_r_20_de),
)
svds_10_v_20 = (
(svd_a_10_sp, svd_a_20_sp),
(svd_r_10_sp, svd_r_20_sp),
(svd_a_10_de, svd_a_20_de),
(svd_r_10_de, svd_r_20_de),
)
svds_sparse_v_dense = (
(svd_a_10_sp, svd_a_10_de),
(svd_a_20_sp, svd_a_20_de),
(svd_r_10_sp, svd_r_10_de),
(svd_r_20_sp, svd_r_20_de),
)
# Assert the 1st component is equal
for svd_10, svd_20 in svds_10_v_20:
assert_array_almost_equal(
svd_10.explained_variance_ratio_,
svd_20.explained_variance_ratio_[:10],
decimal=5,
)
# Assert that 20 components has higher explained variance than 10
for svd_10, svd_20 in svds_10_v_20:
assert_greater(
svd_20.explained_variance_ratio_.sum(),
svd_10.explained_variance_ratio_.sum(),
)
# Assert that all the values are greater than 0
for svd in svds:
assert_array_less(0.0, svd.explained_variance_ratio_)
# Assert that total explained variance is less than 1
for svd in svds:
assert_array_less(svd.explained_variance_ratio_.sum(), 1.0)
# Compare sparse vs. dense
for svd_sparse, svd_dense in svds_sparse_v_dense:
assert_array_almost_equal(svd_sparse.explained_variance_ratio_,
svd_dense.explained_variance_ratio_)
# Test that explained_variance is correct
for svd, transformed in svds_trans:
total_variance = np.var(X.toarray(), axis=0).sum()
variances = np.var(transformed, axis=0)
true_explained_variance_ratio = variances / total_variance
assert_array_almost_equal(
svd.explained_variance_ratio_,
true_explained_variance_ratio,
)
| bsd-3-clause |
rsignell-usgs/PySeidon | pyseidon/tidegaugeClass/plotsTidegauge.py | 2 | 1096 | #!/usr/bin/python2.7
# encoding: utf-8
from __future__ import division
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.tri as Tri
import matplotlib.ticker as ticker
import seaborn
class PlotsTidegauge:
"""'Plots' subset of Tidegauge class gathers plotting functions"""
def __init__(self, variable, debug=False):
self._var = variable
def plot_xy(self, x, y, title=' ', xLabel=' ', yLabel=' '):
"""
Simple X vs Y plot
Inputs:
------
- x = 1D array
- y = 1D array
"""
fig = plt.figure(figsize=(18,10))
plt.rc('font',size='22')
self._fig = plt.plot(x, y, label=title)
scale = 1
ticks = ticker.FuncFormatter(lambda lon, pos: '{0:g}'.format(lon/scale))
plt.ylabel(yLabel)
plt.xlabel(xLabel)
#plt.legend()
plt.show()
#TR_comments: templates
# def whatever(self, debug=False):
# if debug or self._debug:
# print 'Start whatever...'
#
# if debug or self._debug:
# print '...Passed'
| agpl-3.0 |
chrismattmann/tika-similarity | sk_kmeans.py | 2 | 4409 | #!/usr/bin/env python2.7
#
# Licensed to the Apache Software Foundation (ASF) under one or more
# contributor license agreements. See the NOTICE file distributed with
# this work for additional information regarding copyright ownership.
# The ASF licenses this file to You under the Apache License, Version 2.0
# (the "License"); you may not use this file except in compliance with
# the License. You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
#
#
from tika import parser
import pandas as pd
from vector import Vector
from sklearn.cluster import KMeans
import argparse, os, json
def filterFiles(inputDir, acceptTypes):
filename_list = []
for root, dirnames, files in os.walk(inputDir):
dirnames[:] = [d for d in dirnames if not d.startswith('.')]
for filename in files:
if not filename.startswith('.'):
filename_list.append(os.path.join(root, filename))
filename_list = (filename for filename in filename_list if "metadata" in parser.from_file(filename))
if acceptTypes:
filename_list = (filename for filename in filename_list if str(parser.from_file(filename)['metadata']['Content-Type'].encode('utf-8').decode('utf-8')).split('/')[-1] in acceptTypes)
else:
print("Accepting all MIME Types.....")
return filename_list
if __name__ == "__main__":
argParser = argparse.ArgumentParser('k-means Clustering of documents based on metadata values')
argParser.add_argument('--inputDir', required=True, help='path to directory containing files')
argParser.add_argument('--outJSON', required=True, help='/path/to/clusters.json containing k-means cluster assignments')
argParser.add_argument('--Kvalue', help='number of clusters to find')
#argParser.add_argument('--findK', action='store_true', help='find the optimal value of K')
argParser.add_argument('--accept', nargs='+', type=str, help='Optional: compute similarity only on specified IANA MIME Type(s)')
args = argParser.parse_args()
# cluster for a particular value of K
# if args.inputDir and args.outJSON and args.findK:
if args.inputDir and args.outJSON and args.Kvalue:
list_of_points = []
for eachFile in filterFiles(args.inputDir, args.accept):
list_of_points.append(Vector(eachFile, parser.from_file(eachFile)["metadata"]))
list_of_Dicts = (point.features for point in list_of_points)
df = pd.DataFrame(list_of_Dicts)
df = df.fillna(0)
print(df.shape)
kmeans = KMeans(n_clusters=int(args.Kvalue),
init='k-means++',
max_iter=300, # k-means convergence
n_init=10, # find global minima
n_jobs=-2, # parallelize
)
labels = kmeans.fit_predict(df) # unsupervised (X, y=None)
print(labels) # kmeans.labels_
clusters = {}
for i in range(0, len(labels)):
node = { "metadata": json.dumps(list_of_points[i].features),
"name": list_of_points[i].filename.split('/')[-1],
"path": list_of_points[i].filename
}
try:
clusters[str(labels[i])].append(node)
except KeyError:
clusters[str(labels[i])] = []
clusters[str(labels[i])].append(node)
# generate clusters.JSON
with open(args.outJSON, "w") as jsonF:
json_data = {"name": "clusters"}
children = []
for key in clusters:
cluster_children = {"name": "cluster"+key, "children": clusters[key]}
children.append(cluster_children)
json_data["children"] = children
json.dump(json_data, jsonF)
# print matplotlib
# user chooses k => generates k
# find elbow
#kmeans.transform()
# String Length Of Course
# df.to_csv("bashhshs.csv", sep=',')
| apache-2.0 |
dtusar/coco | code-postprocessing/bbob_pproc/compall/pprldmany.py | 3 | 29654 | #! /usr/bin/env python
# -*- coding: utf-8 -*-
"""Generates figure of the bootstrap distribution of ERT.
The main method in this module generates figures of Empirical
Cumulative Distribution Functions of the bootstrap distribution of
the Expected Running Time (ERT) divided by the dimension for many
algorithms.
The outputs show the ECDFs of the running times of the simulated runs
divided by dimension for 50 different targets logarithmically uniformly
distributed in [1e−8, 1e2]. The crosses (×) give the median number of
function evaluations of unsuccessful runs divided by dimension.
**Example**
.. plot::
:width: 50%
import urllib
import tarfile
import glob
from pylab import *
import bbob_pproc as bb
# Collect and unarchive data (3.4MB)
dataurl = 'http://coco.lri.fr/BBOB2009/pythondata/BIPOP-CMA-ES.tar.gz'
filename, headers = urllib.urlretrieve(dataurl)
archivefile = tarfile.open(filename)
archivefile.extractall()
# Empirical cumulative distribution function of bootstrapped ERT figure
ds = bb.load(glob.glob('BBOB2009pythondata/BIPOP-CMA-ES/ppdata_f0*_20.pickle'))
figure()
bb.compall.pprldmany.plot(ds) # must rather call main instead of plot?
bb.compall.pprldmany.beautify()
"""
from __future__ import absolute_import
import os
import warnings
from pdb import set_trace
import numpy as np
import matplotlib.pyplot as plt
from .. import toolsstats, bestalg, genericsettings
from .. import pproc as pp # import dictAlgByDim, dictAlgByFun
from .. import toolsdivers # strip_pathname, str_to_latex
from .. import pprldistr # plotECDF, beautifyECDF
from .. import ppfig # consecutiveNumbers, saveFigure, plotUnifLogXMarkers, logxticks
from .. import pptex # numtotex
displaybest2009 = True
target_values = pp.TargetValues(10**np.arange(2, -8, -0.2)) # possibly changed in config
x_limit = None # not sure whether this is necessary/useful
x_limit_default = 1e7 # better: 10 * genericsettings.evaluation_setting[1], noisy: 1e8, otherwise: 1e7. maximal run length shown
divide_by_dimension = True
annotation_line_end_relative = 1.11 # lines between graph and annotation
annotation_space_end_relative = 1.24 # figure space end relative to x_limit
save_zoom = False # save zoom into left and right part of the figures
perfprofsamplesize = genericsettings.simulated_runlength_bootstrap_sample_size_rld # number of bootstrap samples drawn for each fct+target in the performance profile
dpi_global_var = 100 # 100 ==> 800x600 (~160KB), 120 ==> 960x720 (~200KB), 150 ==> 1200x900 (~300KB) looks ugly in latex
nbperdecade = 1
median_max_evals_marker_format = ['x', 24, 3]
label_fontsize = 18
styles = [d.copy() for d in genericsettings.line_styles] # deep copy
refcolor = 'wheat'
"""color of reference (best) algorithm"""
save_figure = True
close_figure = True
# TODO: update the list below which are not relevant anymore
best = ('AMaLGaM IDEA', 'iAMaLGaM IDEA', 'VNS (Garcia)', 'MA-LS-Chain', 'BIPOP-CMA-ES', 'IPOP-SEP-CMA-ES',
'BFGS', 'NELDER (Han)', 'NELDER (Doe)', 'NEWUOA', 'full NEWUOA', 'GLOBAL', 'MCS (Neum)',
'DIRECT', 'DASA', 'POEMS', 'Cauchy EDA', 'Monte Carlo')
best2 = ('AMaLGaM IDEA', 'iAMaLGaM IDEA', 'VNS (Garcia)', 'MA-LS-Chain', 'BIPOP-CMA-ES', 'IPOP-SEP-CMA-ES', 'BFGS', 'NEWUOA', 'GLOBAL')
eseda = ('AMaLGaM IDEA', 'iAMaLGaM IDEA', 'VNS (Garcia)', 'MA-LS-Chain', 'BIPOP-CMA-ES', 'IPOP-SEP-CMA-ES', '(1+1)-CMA-ES', '(1+1)-ES')
ESs = ('BIPOP-CMA-ES', 'IPOP-SEP-CMA-ES', '(1+1)-CMA-ES', '(1+1)-ES', 'BIPOP-ES')
bestnoisy = ()
bestbest = ('BIPOP-CMA-ES', 'NEWUOA', 'GLOBAL', 'NELDER (Doe)')
nikos = ('AMaLGaM IDEA', 'VNS (Garcia)', 'MA-LS-Chain', 'BIPOP-CMA-ES', '(1+1)-CMA-ES', 'G3-PCX', 'NEWUOA',
'Monte Carlo', 'NELDER (Han)', 'NELDER (Doe)', 'GLOBAL', 'MCS (Neum)')
nikos = ('AMaLGaM IDEA', 'VNS (Garcia)', 'MA-LS-Chain', 'BIPOP-CMA-ES',
'(1+1)-CMA-ES', '(1+1)-ES', 'IPOP-SEP-CMA-ES', 'BIPOP-ES',
'NEWUOA',
'NELDER (Doe)', 'BFGS', 'Monte Carlo')
nikos40D = ('AMaLGaM IDEA', 'iAMaLGaM IDEA', 'BIPOP-CMA-ES',
'(1+1)-CMA-ES', '(1+1)-ES', 'IPOP-SEP-CMA-ES',
'NEWUOA', 'NELDER (Han)', 'BFGS', 'Monte Carlo')
# three groups which include all algorithms:
GA = ('DE-PSO', '(1+1)-ES', 'PSO_Bounds', 'DASA', 'G3-PCX', 'simple GA', 'POEMS', 'Monte Carlo') # 7+1
classics = ('BFGS', 'NELDER (Han)', 'NELDER (Doe)', 'NEWUOA', 'full NEWUOA', 'DIRECT', 'LSfminbnd',
'LSstep', 'Rosenbrock', 'GLOBAL', 'SNOBFIT', 'MCS (Neum)', 'adaptive SPSA', 'Monte Carlo') # 13+1
EDA = ('BIPOP-CMA-ES', '(1+1)-CMA-ES', 'VNS (Garcia)', 'EDA-PSO', 'IPOP-SEP-CMA-ES', 'AMaLGaM IDEA',
'iAMaLGaM IDEA', 'Cauchy EDA', 'BayEDAcG', 'MA-LS-Chain', 'Monte Carlo') # 10+1
# groups according to the talks
petr = ('DIRECT', 'LSfminbnd', 'LSstep', 'Rosenbrock', 'G3-PCX', 'Cauchy EDA', 'Monte Carlo')
TAO = ('BFGS', 'NELDER (Han)', 'NEWUOA', 'full NEWUOA', 'BIPOP-CMA-ES', 'IPOP-SEP-CMA-ES',
'(1+1)-CMA-ES', '(1+1)-ES', 'simple GA', 'Monte Carlo')
TAOp = TAO + ('NELDER (Doe)',)
MC = ('Monte Carlo',)
third = ('POEMS', 'VNS (Garcia)', 'DE-PSO', 'EDA-PSO', 'PSO_Bounds', 'PSO', 'AMaLGaM IDEA', 'iAMaLGaM IDEA',
'MA-LS-Chain', 'DASA', 'BayEDAcG')
funi = [1,2] + range(5, 15) # 2 is paired Ellipsoid
funilipschitz = [1] + [5,6] + range(8,13) + [14] # + [13] #13=sharp ridge, 7=step-ellipsoid
fmulti = [3, 4] + range(15,25) # 3 = paired Rastrigin
funisep = [1,2,5]
# input parameter settings
show_algorithms = eseda + ('BFGS',) # ()==all
#show_algorithms = ('IPOP-SEP-CMA-ES', 'IPOP-CMA-ES', 'BIPOP-CMA-ES',)
#show_algorithms = ('IPOP-SEP-CMA-ES', 'IPOP-CMA-ES', 'BIPOP-CMA-ES',
#'avg NEWUOA', 'NEWUOA', 'full NEWUOA', 'BFGS', 'MCS (Neum)', 'GLOBAL', 'NELDER (Han)',
#'NELDER (Doe)', 'Monte Carlo') # ()==all
show_algorithms = () # could be one of the list above
function_IDs = ()
function_IDs = range(1,200) # sep ros high mul mulw == 1, 6, 10, 15, 20, 101, 107, 122,
#function_IDs = range(101,199) # sep ros high mul mulw == 1, 6, 10, 15, 20, 101, 107, 122,
#function_IDs = fmulti # funi fmulti # range(103, 131, 3) # displayed functions
#function_IDs = [1,2,3,4,5] # separable functions
#function_IDs = [6,7,8,9] # moderate functions
#function_IDs = [10,11,12,13,14] # ill-conditioned functions
#function_IDs = [15,16,17,18,19] # multi-modal functions
#function_IDs = [20,21,22,23,24] # weak structure functions
#function_IDs = range(101,131) # noisy testbed
#function_IDs = range(101,106+1) # moderate noise
#function_IDs = range(107,130+1) # severe noise
#function_IDs = range(101,130+1, 3) # gauss noise
#function_IDs = range(102,130+1, 3) # unif noise
#function_IDs = range(103,130+1, 3) # cauchy noise
# function_IDs = range(15,25) # multimodal nonseparable
#'-' solid line style
#'--' dashed line style
#'-.' dash-dot line style
#':' dotted line style
#'.' point marker
#',' pixel marker
#'o' circle marker
#'v' triangle_down marker
#'^' triangle_up marker
#'<' triangle_left marker
#'>' triangle_right marker
#'1' tri_down marker
#'2' tri_up marker
#'3' tri_left marker
#'4' tri_right marker
#'s' square marker
#'p' pentagon marker
#'*' star marker
#'h' hexagon1 marker
#'H' hexagon2 marker
#'+' plus marker
#'x' x marker
#'D' diamond marker
#'d' thin_diamond marker
#'|' vline marker
#'_' hline marker
def plt_plot(*args, **kwargs):
return plt.plot(*args, clip_on=False, **kwargs)
def beautify():
"""Customize figure presentation."""
#plt.xscale('log') # Does not work with matplotlib 0.91.2
a = plt.gca()
a.set_xscale('log')
#Tick label handling
plt.xlim(xmin=1e-0)
global divide_by_dimension
if divide_by_dimension:
plt.xlabel('log10 of (# f-evals / dimension)', fontsize=label_fontsize)
else:
plt.xlabel('log10 of # f-evals', fontsize=label_fontsize)
plt.ylabel('Proportion of function+target pairs', fontsize=label_fontsize)
ppfig.logxticks()
pprldistr.beautifyECDF()
def plotdata(data, maxval=None, maxevals=None, CrE=0., **kwargs):
"""Draw a normalized ECDF. What means normalized?
:param seq data: data set, a 1-D ndarray of runlengths
:param float maxval: right-most value to be displayed, will use the
largest non-inf, non-nan value in data if not
provided
:param seq maxevals: if provided, will plot the median of this
sequence as a single cross marker
:param float CrE: Crafting effort the data will be multiplied by
the exponential of this value.
:param kwargs: optional arguments provided to plot function.
"""
#Expect data to be a ndarray.
x = data[np.isnan(data)==False] # Take away the nans
nn = len(x)
x = x[np.isinf(x)==False] # Take away the infs
n = len(x)
x = np.exp(CrE) * x # correction by crafting effort CrE
if n == 0:
#res = plt.plot((1., ), (0., ), **kwargs)
res = pprldistr.plotECDF(np.array((1., )), n=np.inf, **kwargs)
else:
dictx = {} # number of appearances of each value in x
for i in x:
dictx[i] = dictx.get(i, 0) + 1
x = np.array(sorted(dictx)) # x is not a multiset anymore
y = np.cumsum(list(dictx[i] for i in x)) # cumsum of size of y-steps (nb of appearences)
idx = sum(x <= x_limit**annotation_space_end_relative) - 1
y_last, x_last = y[idx] / float(nn), x[idx]
if maxval is None:
maxval = max(x)
end = np.sum(x <= maxval)
x = x[:end]
y = y[:end]
try: # plot the very last point outside of the "normal" plotting area
c = kwargs['color']
plt_plot([x_last] * 2, [y_last] * 2, '.', color=c, markeredgecolor=c)
except:
pass
x2 = np.hstack([np.repeat(x, 2), maxval]) # repeat x-values for each step in the cdf
y2 = np.hstack([0.0, np.repeat(y / float(nn), 2)])
res = ppfig.plotUnifLogXMarkers(x2, y2, nbperdecade * 3 / np.log10(maxval),
logscale=False, clip_on=False, **kwargs)
# res = plotUnifLogXMarkers(x2, y2, nbperdecade, logscale=False, **kwargs)
if maxevals: # Should cover the case where maxevals is None or empty
x3 = np.median(maxevals)
if (x3 <= maxval and
# np.any(x2 <= x3) and # maxval < median(maxevals)
not plt.getp(res[-1], 'label').startswith('best')
): # TODO: HACK for not considering the best 2009 line
try:
y3 = y2[x2<=x3][-1] # find right y-value for x3==median(maxevals)
except IndexError: # median(maxevals) is smaller than any data, can only happen because of CrE?
y3 = y2[0]
h = plt.plot((x3,), (y3,),
marker=median_max_evals_marker_format[0],
markersize=median_max_evals_marker_format[1],
markeredgewidth=median_max_evals_marker_format[2],
# marker='x', markersize=24, markeredgewidth=3,
markeredgecolor=plt.getp(res[0], 'color'),
ls=plt.getp(res[0], 'ls'),
color=plt.getp(res[0], 'color'))
h.extend(res)
res = h # so the last element in res still has the label.
# Only take sequences for x and y!
return res
def plotLegend(handles, maxval):
"""Display right-side legend.
:param float maxval: rightmost x boundary
:returns: list of (ordered) labels and handles.
The figure is stopped at maxval (upper x-bound), and the graphs in
the figure are prolonged with straight lines to the right to connect
with labels of the graphs (uniformly spread out vertically). The
order of the graphs at the upper x-bound line give the order of the
labels, in case of ties, the best is the graph for which the x-value
of the first step (from the right) is smallest.
The annotation string is stripped from preceeding pathnames.
"""
reslabels = []
reshandles = []
ys = {}
lh = 0
for h in handles:
x2 = []
y2 = []
for i in h:
x2.append(plt.getp(i, "xdata"))
y2.append(plt.getp(i, "ydata"))
x2 = np.array(np.hstack(x2))
y2 = np.array(np.hstack(y2))
tmp = np.argsort(x2)
x2 = x2[tmp]
y2 = y2[tmp]
h = h[-1] # we expect the label to be in the last element of h
tmp = (x2 <= maxval)
try:
x2bis = x2[y2 < y2[tmp][-1]][-1]
except IndexError: # there is no data with a y smaller than max(y)
x2bis = 0.
ys.setdefault(y2[tmp][-1], {}).setdefault(x2bis, []).append(h)
lh += 1
if len(show_algorithms) > 0:
lh = min(lh, len(show_algorithms))
if lh <= 1:
lh = 2
fontsize = genericsettings.minmax_algorithm_fontsize[0] + np.min((1, np.exp(9-lh))) * (
genericsettings.minmax_algorithm_fontsize[-1] - genericsettings.minmax_algorithm_fontsize[0])
i = 0 # loop over the elements of ys
for j in sorted(ys.keys()):
for k in reversed(sorted(ys[j].keys())):
#enforce best ever comes last in case of equality
tmp = []
for h in ys[j][k]:
if plt.getp(h, 'label') == 'best 2009':
tmp.insert(0, h)
else:
tmp.append(h)
tmp.reverse()
ys[j][k] = tmp
for h in ys[j][k]:
if (not plt.getp(h, 'label').startswith('_line') and
(len(show_algorithms) == 0 or
plt.getp(h, 'label') in show_algorithms)):
y = 0.02 + i * 0.96/(lh-1)
tmp = {}
for attr in ('lw', 'ls', 'marker',
'markeredgewidth', 'markerfacecolor',
'markeredgecolor', 'markersize', 'zorder'):
tmp[attr] = plt.getp(h, attr)
legx = maxval**annotation_line_end_relative
if 'marker' in attr:
legx = maxval**annotation_line_end_relative
# reshandles.extend(plt_plot((maxval, legx), (j, y),
reshandles.extend(plt_plot((maxval, legx), (j, y),
color=plt.getp(h, 'markeredgecolor'), **tmp))
reshandles.append(
plt.text(maxval**(0.02 + annotation_line_end_relative), y,
toolsdivers.str_to_latex(toolsdivers.strip_pathname1(plt.getp(h, 'label'))),
horizontalalignment="left",
verticalalignment="center", size=fontsize))
reslabels.append(plt.getp(h, 'label'))
#set_trace()
i += 1
#plt.axvline(x=maxval, color='k') # Not as efficient?
reshandles.append(plt_plot((maxval, maxval), (0., 1.), color='k'))
reslabels.reverse()
plt.xlim(xmax=maxval**annotation_space_end_relative)
return reslabels, reshandles
def plot(dsList, targets=None, craftingeffort=0., **kwargs):
"""This function is obsolete?
Generates a graph of the run length distribution of an algorithm.
We display the empirical cumulative distribution function ECDF of
the bootstrapped distribution of the runlength for an algorithm
(in number of function evaluations) to reach the target functions
value :py:data:`targets`.
:param DataSetList dsList: data set for one algorithm
:param seq targets: target function values
:param float crafting effort: the data will be multiplied by the
exponential of this value
:param dict kwargs: additional parameters provided to plot function.
:returns: handles
"""
if targets is None:
targets = target_values # set above or in config.py
try:
if np.min(targets) >= 1:
ValueError('smallest target f-value is not smaller than one, use ``pproc.TargetValues(targets)`` to prevent this error')
targets = pp.TargetValues(targets)
except TypeError:
pass
res = []
assert len(pp.DataSetList(dsList).dictByDim()) == 1 # We never integrate over dimensions...
data = []
maxevals = []
for entry in dsList:
for t in targets((entry.funcId, entry.dim)):
divisor = entry.dim if divide_by_dimension else 1
x = [np.inf] * perfprofsamplesize
runlengthunsucc = []
evals = entry.detEvals([t])[0]
runlengthsucc = evals[np.isnan(evals) == False] / divisor
runlengthunsucc = entry.maxevals[np.isnan(evals)] / divisor
if len(runlengthsucc) > 0:
x = toolsstats.drawSP(runlengthsucc, runlengthunsucc,
percentiles=[50],
samplesize=perfprofsamplesize)[1]
data.extend(x)
maxevals.extend(runlengthunsucc)
# Display data
data = np.array(data)
data = data[np.isnan(data)==False] # Take away the nans
n = len(data)
data = data[np.isinf(data)==False] # Take away the infs
# data = data[data <= maxval] # Take away rightmost data
data = np.exp(craftingeffort) * data # correction by crafting effort CrE
if len(data) == 0: # data is empty.
res = pprldistr.plotECDF(np.array((1., )), n=np.inf, **kwargs)
else:
res = pprldistr.plotECDF(np.array(data), n=n, **kwargs)
#plotdata(np.array(data), x_limit, maxevals,
# CrE=0., **kwargs)
if maxevals: # Should cover the case where maxevals is None or empty
x3 = np.median(maxevals)
if np.any(data > x3):
y3 = float(np.sum(data <= x3)) / n
h = plt_plot((x3,), (y3,), marker='x', markersize=24, markeredgewidth=3,
markeredgecolor=plt.getp(res[0], 'color'),
ls='', color=plt.getp(res[0], 'color'))
h.extend(res)
res = h # so the last element in res still has the label.
return res
def all_single_functions(dictAlg, sortedAlgs=None, outputdir='.',
verbose=0):
dictFG = pp.dictAlgByFun(dictAlg)
for fg, tmpdictAlg in dictFG.iteritems():
dictDim = pp.dictAlgByDim(tmpdictAlg)
for d, entries in dictDim.iteritems():
single_fct_output_dir = (outputdir.rstrip(os.sep) + os.sep +
'pprldmany-single-functions'
# + os.sep + ('f%03d' % fg)
)
if not os.path.exists(single_fct_output_dir):
os.makedirs(single_fct_output_dir)
main(entries,
order=sortedAlgs,
outputdir=single_fct_output_dir,
info=('f%03d_%02dD' % (fg, d)),
verbose=verbose)
def main(dictAlg, isBiobjective, order=None, outputdir='.', info='default',
dimension=None, verbose=True):
"""Generates a figure showing the performance of algorithms.
From a dictionary of :py:class:`DataSetList` sorted by algorithms,
generates the cumulative distribution function of the bootstrap
distribution of ERT for algorithms on multiple functions for
multiple targets altogether.
:param dict dictAlg: dictionary of :py:class:`DataSetList` instances
one instance is equivalent to one algorithm,
:param list targets: target function values
:param list order: sorted list of keys to dictAlg for plotting order
:param str outputdir: output directory
:param str info: output file name suffix
:param bool verbose: controls verbosity
"""
global x_limit # late assignment of default, because it can be set to None in config
global divide_by_dimension # not fully implemented/tested yet
if 'x_limit' not in globals() or x_limit is None:
x_limit = x_limit_default
tmp = pp.dictAlgByDim(dictAlg)
# tmp = pp.DictAlg(dictAlg).by_dim()
if len(tmp) != 1 and dimension is None:
raise ValueError('We never integrate over dimension.')
if dimension is not None:
if dimension not in tmp.keys():
raise ValueError('dimension %d not in dictAlg dimensions %s'
% (dimension, str(tmp.keys())))
tmp = {dimension: tmp[dimension]}
dim = tmp.keys()[0]
divisor = dim if divide_by_dimension else 1
algorithms_with_data = [a for a in dictAlg.keys() if dictAlg[a] != []]
dictFunc = pp.dictAlgByFun(dictAlg)
# Collect data
# Crafting effort correction: should we consider any?
CrEperAlg = {}
for alg in algorithms_with_data:
CrE = 0.
if 1 < 3 and dictAlg[alg][0].algId == 'GLOBAL':
tmp = dictAlg[alg].dictByNoise()
assert len(tmp.keys()) == 1
if tmp.keys()[0] == 'noiselessall':
CrE = 0.5117
elif tmp.keys()[0] == 'nzall':
CrE = 0.6572
CrEperAlg[alg] = CrE
if CrE != 0.0:
print 'Crafting effort for', alg, 'is', CrE
dictData = {} # list of (ert per function) per algorithm
dictMaxEvals = {} # list of (maxevals per function) per algorithm
bestERT = [] # best ert per function
# funcsolved = [set()] * len(targets) # number of functions solved per target
xbest2009 = []
maxevalsbest2009 = []
for f, dictAlgperFunc in dictFunc.iteritems():
if function_IDs and f not in function_IDs:
continue
# print target_values((f, dim))
for j, t in enumerate(target_values((f, dim))):
# for j, t in enumerate(genericsettings.current_testbed.ecdf_target_values(1e2, f)):
# funcsolved[j].add(f)
for alg in algorithms_with_data:
x = [np.inf] * perfprofsamplesize
runlengthunsucc = []
try:
entry = dictAlgperFunc[alg][0] # one element per fun and per dim.
evals = entry.detEvals([t])[0]
assert entry.dim == dim
runlengthsucc = evals[np.isnan(evals) == False] / divisor
runlengthunsucc = entry.maxevals[np.isnan(evals)] / divisor
if len(runlengthsucc) > 0:
x = toolsstats.drawSP(runlengthsucc, runlengthunsucc,
percentiles=[50],
samplesize=perfprofsamplesize)[1]
except (KeyError, IndexError):
#set_trace()
warntxt = ('Data for algorithm %s on function %d in %d-D '
% (alg, f, dim)
+ 'are missing.\n')
warnings.warn(warntxt)
dictData.setdefault(alg, []).extend(x)
dictMaxEvals.setdefault(alg, []).extend(runlengthunsucc)
displaybest2009 = not isBiobjective #disabled until we find the bug
if displaybest2009:
#set_trace()
bestalgentries = bestalg.loadBestAlgorithm(isBiobjective)
bestalgentry = bestalgentries[(dim, f)]
bestalgevals = bestalgentry.detEvals(target_values((f, dim)))
# print bestalgevals
for j in range(len(bestalgevals[0])):
if bestalgevals[1][j]:
evals = bestalgevals[0][j]
#set_trace()
assert dim == bestalgentry.dim
runlengthsucc = evals[np.isnan(evals) == False] / divisor
runlengthunsucc = bestalgentry.maxevals[bestalgevals[1][j]][np.isnan(evals)] / divisor
x = toolsstats.drawSP(runlengthsucc, runlengthunsucc,
percentiles=[50],
samplesize=perfprofsamplesize)[1]
else:
x = perfprofsamplesize * [np.inf]
runlengthunsucc = []
xbest2009.extend(x)
maxevalsbest2009.extend(runlengthunsucc)
if order is None:
order = dictData.keys()
# Display data
lines = []
if displaybest2009:
args = {'ls': '-', 'linewidth': 6, 'marker': 'D', 'markersize': 11.,
'markeredgewidth': 1.5, 'markerfacecolor': refcolor,
'markeredgecolor': refcolor, 'color': refcolor,
'label': 'best 2009', 'zorder': -1}
lines.append(plotdata(np.array(xbest2009), x_limit, maxevalsbest2009,
CrE = 0., **args))
def algname_to_label(algname, dirname=None):
"""to be extended to become generally useful"""
if isinstance(algname, (tuple, list)): # not sure this is needed
return ' '.join([str(name) for name in algname])
return str(algname)
for i, alg in enumerate(order):
try:
data = dictData[alg]
maxevals = dictMaxEvals[alg]
except KeyError:
continue
args = styles[(i) % len(styles)]
args['linewidth'] = 1.5
args['markersize'] = 12.
args['markeredgewidth'] = 1.5
args['markerfacecolor'] = 'None'
args['markeredgecolor'] = args['color']
args['label'] = algname_to_label(alg)
#args['markevery'] = perfprofsamplesize # option available in latest version of matplotlib
#elif len(show_algorithms) > 0:
#args['color'] = 'wheat'
#args['ls'] = '-'
#args['zorder'] = -1
# plotdata calls pprldistr.plotECDF which calls ppfig.plotUnifLog... which does the work
lines.append(plotdata(np.array(data), x_limit, maxevals,
CrE=CrEperAlg[alg], **args))
labels, handles = plotLegend(lines, x_limit)
if True: # isLateXLeg:
fileName = os.path.join(outputdir,'pprldmany_%s.tex' % (info))
with open(fileName, 'w') as f:
f.write(r'\providecommand{\nperfprof}{7}')
algtocommand = {} # latex commands
for i, alg in enumerate(order):
tmp = r'\alg%sperfprof' % pptex.numtotext(i)
f.write(r'\providecommand{%s}{\StrLeft{%s}{\nperfprof}}' %
(tmp, toolsdivers.str_to_latex(
toolsdivers.strip_pathname2(algname_to_label(alg)))))
algtocommand[algname_to_label(alg)] = tmp
if displaybest2009:
tmp = r'\algzeroperfprof'
f.write(r'\providecommand{%s}{best 2009}' % (tmp))
algtocommand['best 2009'] = tmp
commandnames = []
for label in labels:
commandnames.append(algtocommand[label])
# f.write(headleg)
if len(order) > 28: # latex sidepanel won't work well for more than 25 algorithms, but original labels are also clipped
f.write(r'\providecommand{\perfprofsidepanel}{\mbox{%s}\vfill\mbox{%s}}'
% (commandnames[0], commandnames[-1]))
else:
fontsize_command = r'\tiny{}' if len(order) > 19 else ''
f.write(r'\providecommand{\perfprofsidepanel}{{%s\mbox{%s}' %
(fontsize_command, commandnames[0])) # TODO: check len(labels) > 0
for i in range(1, len(labels)):
f.write('\n' + r'\vfill \mbox{%s}' % commandnames[i])
f.write('}}\n')
# f.write(footleg)
if verbose:
print 'Wrote right-hand legend in %s' % fileName
figureName = os.path.join(outputdir,'pprldmany_%s' % (info))
#beautify(figureName, funcsolved, x_limit*x_annote_factor, False, fileFormat=figformat)
beautify()
text = ppfig.consecutiveNumbers(sorted(dictFunc.keys()), 'f')
text += ',%d-D' % dim # TODO: this is strange when different dimensions are plotted
plt.text(0.01, 0.98, text, horizontalalignment="left",
verticalalignment="top", transform=plt.gca().transAxes)
if len(dictFunc) == 1:
plt.title(' '.join((str(dictFunc.keys()[0]),
genericsettings.current_testbed.short_names[dictFunc.keys()[0]])))
a = plt.gca()
plt.xlim(xmin=1e-0, xmax=x_limit**annotation_space_end_relative)
xticks, labels = plt.xticks()
tmp = []
for i in xticks:
tmp.append('%d' % round(np.log10(i)))
a.set_xticklabels(tmp)
if save_figure:
ppfig.saveFigure(figureName, verbose=verbose)
if len(dictFunc) == 1:
ppfig.save_single_functions_html(
os.path.join(outputdir, 'pprldmany'),
'', # algorithms names are clearly visible in the figure
add_to_names='_%02dD' %(dim),
algorithmCount=ppfig.AlgorithmCount.NON_SPECIFIED
)
if close_figure:
plt.close()
# TODO: should return status or sthg
if __name__ == "__main__":
# should become a test case
import sys
import bbob_pproc
sys.path.append('.')
| bsd-3-clause |